Physical Biochemistry
Our research is focused on
- Ribosome function and dynamics
- Regulation and fidelity of translation
- Ribosome-catalyzed reactions
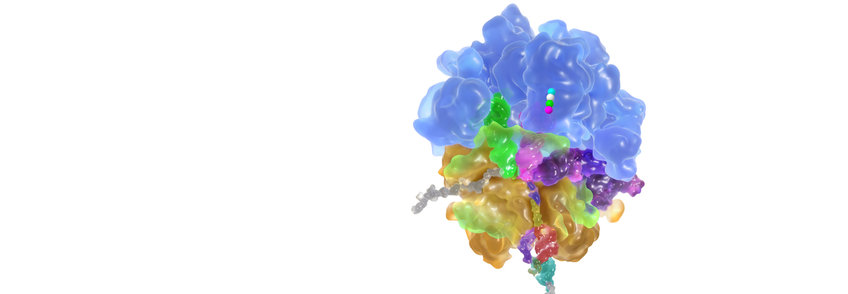
Protein synthesis from amino acids in the cell is performed on ribosomes, large ribonucleoprotein particles that consist of several RNA molecules and over 50 proteins. The ribosome is a molecular machine that selects its substrates, aminoacyl-tRNAs, very rapidly and accurately and catalyses the synthesis of peptides from amino acids. Recently, enormous progress has been made in understanding the ribosome by analyzing its functions using biophysical techniques and solving ribosome structures by crystallography and cryoelectron microscopy. The functional and structural basis for the fidelity with which mRNA sequences are translated into polypeptide sequences is understood in great detail, and the main factors contributing to the catalysis of peptide-bond formation are identified. In addition, much has been learned about the mechanisms by which antibiotics inhibit ribosome function.
The active site for peptide bond formation – the peptidyl transferase center – is composed of ribosomal RNA, and thus the ribosome is the largest known RNA catalyst, and the only natural ribozyme that has polymerase activity. The ribosome employs entropic catalysis to accelerate peptide bond formation by positioning substrates, reorganizing water in the active site, and providing an electrostatic network that stabilizes reaction intermediates. Proton transfer during the reaction appears to be promoted by a concerted shuttle mechanism that involves ribose hydroxyl groups on the tRNA substrate.
Aminoacyl-tRNA is delivered to the ribosome in a ternary complex with elongation factor Tu (EF-Tu) and GTP. The stepwise movement of aminoacyl-tRNA from EF-Tu into the ribosomal A site entails a number of intermediates. We have discovered the induced-fit mechanism of tRNA selection on the ribosome. Kinetic measurements showed that the ribosome not only bound correct substrates more tightly than the incorrect ones (stability discrimination), but also accelerated the rate of forward reactions for correct, rather than for incorrect tRNA (induced fit). This entirely new concept provided the basis for the interpretation of the interactions and conformational changes in the crystal structured of ribosomes and cryo-EM reconstructions of ribosome complexes. Codon reading by the anticodon of tRNA is controlled by a network of ribosome contacts that are specific for each position of the codon-anticodon duplex and involve A-minor RNA interactions. Rapid and accurate tRNA selection is accomplished by switching the conformation of the decoding site between accepting and rejecting mode, regardless of the thermodynamic stability of the respective codon-anticodon complexes or their interactions at the decoding site. The forward reactions are particularly sensitive to mismatches and determine the variations in the extent of misreading of near-cognate codons, both during initial selection and proofreading.
Among the most important unresolved questions is the role of structural dynamics in ribosome function. The communication between the functional centers of the ribosome is known to be crucial, but there are only vague ideas as to how this may take place. The activation of the GTPase of elongation factor (EF)-Tu is a key step in selection of aminoacyl tRNAs by the ribosome. It is triggered by events on the small subunit, but the GTP-binding site of EF-Tu associates with the large subunit, and the way the signal is transmitted within the ribosome remains unknown. The mechanism of the translocation step, i.e. the movement of tRNAs and mRNA through the ribosome, remains a major challenge. EF-G accelerates translocation by using the energy of GTP hydrolysis to drive translocation which resembles the way motor proteins work; however, the structural basis for the movement and its biophysical characteristics are not known. Finally, incorporation of unusual amino acids, such as selenocysteine, requires highly specialized machinery for delivery; very little is known about the molecular mechanism of this process. None of these problems can be solved without using a combination of techniques from Biochemistry, Structural Biology and Physical Biochemistry and developing new approaches to structure, function, and dynamics of the translational apparatus.
In a broader context, the ribosome can serve as a well-characterized model of large macromolecular assemblies. Using the biophysical approaches devised for the ribosome, it should be possible to obtain information for even larger and more complex macromolecular assemblies. Developing of highly efficient and controlled ribosome translation systems on a highly sophisticated technological level is important for production of proteins with desired properties for purposes of proteomics and high-throughput structural studies emerging in the post-genomic era. The translational apparatus is a major target for antibiotics. Better understanding of the mechanisms of antibiotic action, resistance mechanisms and the interplay between resistance and bacterial fitness using systems biology will be increasingly important for developing new antimicrobials and combating the major infectious diseases.


