Ribosome Dynamics
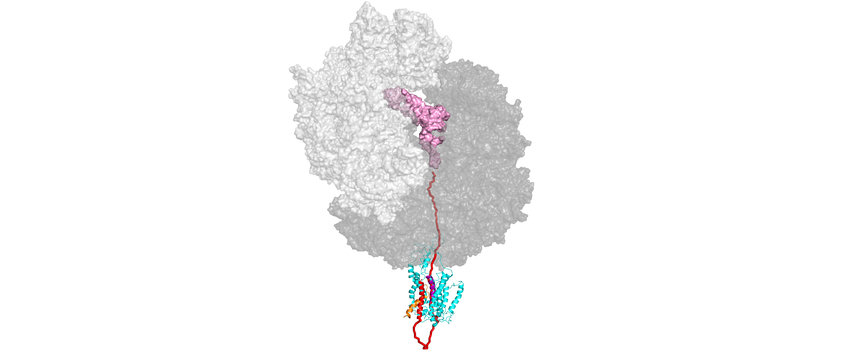
Research interests of the group are aimed at various aspects of ribosomal protein synthesis (see figure), focusing in particular on the biogenesis of integral membrane proteins in bacteria.

In one project we are examining the function of the signal recognition particle (SRP) in the biosynthesis of membrane proteins. SRP binds to ribosomes translating membrane proteins, recognizing a hydrophobic signal sequence (highlighted in red in the figure) that is part of the nascent peptide (light blue or black). By an interaction with the SRP receptor (FtsY) those ribosomes are targeted to the translocation pore (translocon) in the membrane, forming a quaternary transfer complex. We are interested in the dynamics of that complex, in particular the dynamics of the translocon, that allows membrane proteins to insert into the membrane in a specific topology cotranslationally.
In a related project we are studying the interplay between various ribosome-associated protein biogenesis factors, including SRP, the chaperone trigger factor (TF) as well as the modifying enzymes peptide deformylase (PDF) and methionine aminopeptidase (MAP). We use methods of biochemistry and molecular biology, as well as biophysical techniques, such as fluorescence, including single-molecule fluorescence, rapid kinetics (stopped flow, quench flow), and others.
