Fidelity of protein synthesis in vivo
Proteins are versatile macromolecules that fulfil a wide variety of tasks in the cell, such as scaffolding functions, catalysis of chemical reactions or signal transduction. Proteins are synthesized from amino acids by ribosomes according to blueprints encoded in the DNA. Errors during protein synthesis, such as the incorporation of the wrong amino acids, can lead to inactive or misfolded proteins. The accuracy of protein synthesis therefore determines the quality of the proteome and is crucial for the fitness of all cells and organisms. While translation errors in human cells often lead to disease and cellular ageing, errors can also be deliberately provoked by antibiotics to fight pathogenic bacteria and viruses. The aim of our research is to understand the accuracy of protein synthesis by the ribosome, to characterize the errors that occur in the process and the consequences that these errors have in the cell. For this purpose, we use and further develop the analytical method of quantitative mass spectrometry. This technique allows us to determine the frequency of various error types in cellular proteins and correlate them with the associated stress responses.
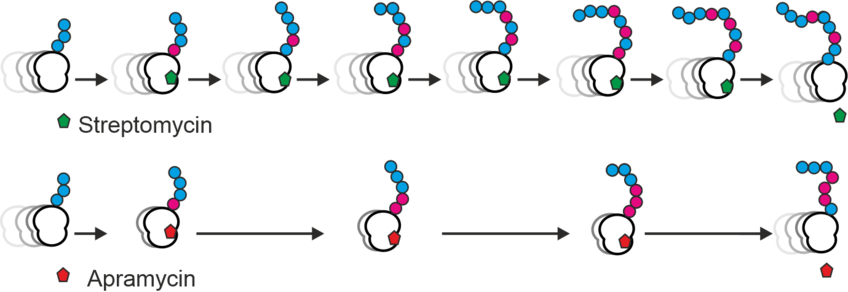
A focus of our research is the elucidation of the mechanism of action of aminoglycosides, an important class of antibiotics. Aminoglycosides kill bacteria by disrupting ribosome function during protein synthesis by causing high levels of errors. We aim to understand how aminoglycosides enter the cell, which types and levels of errors they induce, what the consequences of those errors are and why they lead to cell death.