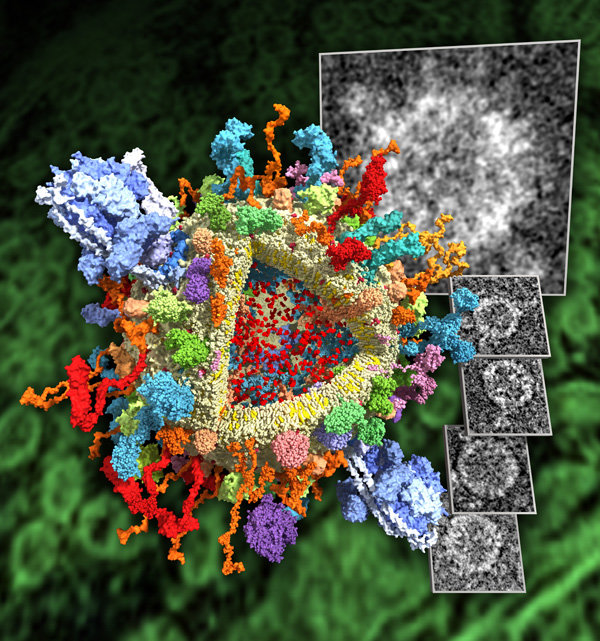
Providing scientifically meaningful illustrations that at the same time captivate the observer is a challenge of its own. Here are some attempts.

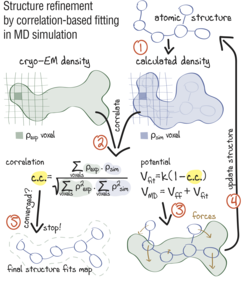
Approach: using correlation-based refinement in MD simulation to steer the atomic positions of a macromolecule such that they optimally fit a cryo-EM map. The molecule is subjected to a global biasing potential Vfit in addition to the MD force field Vff. The forces resulting from Vfit act on every atom to enhance the real-space correlation coefficient 𝑐.𝑐. between the cryo-EM density (green) and the density calculated from the current atomic positions (blue). The first step (1) is to generate a simulated density by convoluting the atomic positions with a three-dimensional Gaussian function of width 𝜎 (Orzechowski and Tama, 2008). The two maps are correlated (2), and the biasing forces are calculated. These forces are then added to the standard MD force field (3), and new atomic positions are evaluated (4). Steps (1–4) are repeated, yielding a structure that correlates better with the cryo-EM map than the starting structure.
Sketch made with Adobe Illustrator.
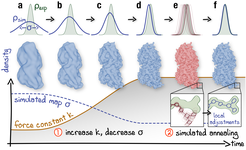
Schematic representation of the proposed continuous refinement protocol: (1) a low temperature optimization phase, where Vfit is monotonously increased by increasing the force constant 𝑘 (columns a–d), followed by (2) simulated annealing (columns e, f). The local effect of the protocol is exemplified in the upper row for a one-dimensional single-atom case. Simulated densities shown in the middle row were generated using the atomic structure of a tubulin dimer (PDB ID: 3JAT; Zhang et al. (2015)). Sketch made with Illustrator.