Ligand Release Pathways and Discrimination in Bombyx mori Pheromone Binding Protein
Frauke Gräter and Helmut Grubmüller
Collaborating groups: Walter Leal (University of California-Davis) and Karl-Ernst Kaissling (MPI Seewiesen)
Supported by: Volkswagen Foundation and Boehringer-Ingelheim-Fonds.
The olfactory system of insects is one of the best biosensors known. It is both extraordinarily selective and sensitive to the female sex pheromone bombykol. To reach the pheromone receptor at the neuronal membrane the hydrophobic bombykol needs to be tranferred through the sensillar lymph of the olfactory hair. This transfer is assisted by a carrier, the pheromone-binding protein of Bombyx mori (BmorPBP), which binds the pheromone molecule, carries it to the receptor, and releases it there. BmorPBP completely encapsulates the ligand and the crystal structure does not suggest any obvious exit/entrance gate for bombykol. In a collaboration with W. Leal (University of California-Davis) and K.-E. Kaissling (MPI Seewiesen), we have shown by using a combination of complementary simulation techniques that, unexpectedly, two binding pathways exist on opposite ends of the binding protein, thus enhancing ligand binding kinetics at the diffusion limit.
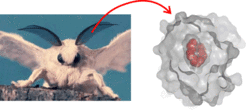
Figure 1: Left: The silk moth Bombyx mori. The male detects minute amounts of the sex pheromone bombykol in the air by means of the olfactory hairs at the antennae. Right: The pheromone binding protein (gray surface) completely encapsulates bombykol (red spheres) in the central binding cavity.
The male silk moth Bombyx mori can detect very low quantities of the sex pheromone bombykol emitted by the female, and at the same time distinguishes bombykol from many other, often similar, volatile compounds in the air. A first step of olfaction is the tranfer of bombykol to the pheromone receptor at the neuronal membrane via the pheromone-binding protein (BmorPBP). Highly efficient uptake and release kinetics of bombykol at the pheromone binding protein is essential for olfactory function. However, BmorPBP features a central hydrophobic binding cavity for bombykol, completely encapsulating the ligand (Figure 1, right) and the exit/entrance gate for bombykol is not known. Yet, the ligand has to enter and exit the cavity fast and reversibly.
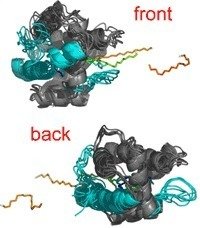
A major step in conceiving the function of BmorPBP is to understand the molecular mechanism of pheromone binding and release. To this end, we examined the mechanism of ligand release from BmorPBP by means of different Molecular Dynamics simulation techniques [1].
Unexpectedly, two binding pathways were found to exist on opposite ends of the binding protein, which both seem to have comparingly low activation barriers. We confirmed our findings by additional force-probe Molecular Dynamics simulations using pulling forces along different chosen directions to probe release pathways. Again, front and back release involve similar forces and free energies. In addition, the molecular mechanisms itselves obtained from Essential Dynamics and force-probe MD simulations were found to be very alike.
Physiological relevance of two ligand entrance/exit pathways: It might render the pheromone binding protein robust towards point mutations. It might also enhance ligand uptake, which here is probably not assisted by electrostatic steering given the largely non-polarity of bombykol, by providing two entrance gates, thereby increasing the chance of ligand uptake upon diffusional encounters of BmorPBP and bombykol.
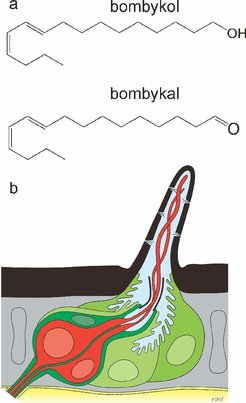
Bombykol and Bombykal: Another question regarding the mode of function of pheromone-binding proteins concerns their contribution to the extraordinary selectivity of the olfactory system of insects. The pheromone of Bombyx mori consists of two components, bombykol and bombykal, the latter acting as an antagonist. The pheromone binding protein was suggested to play the role of a pre-filter preferentially or exclusively carrying bombykol to the receptor. We addressed this question by assessing the difference in binding affinity of the pheromone-binding protein to bombykol and bombykal using free energy perturbation. Surprisingly, we find no difference in the binding affinity for the two pheromone components, ruling out a contribution of the pheromone binding protein to the selectivity of the olfactory system for these two chemically highly similar compounds [2]. Our result was confirmed by additional docking studies and experimental binding assays carried out in the lab of Walter Leal.


