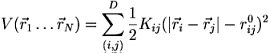FRETsg 1.0 - FRET Structure Generator
Financial Support: Volkswagen Foundation
FRETsg is a tool for a fast qualitative analysis of multiple FRET (fluorescence resonance energy transfer) experiments. FRET experiments principally yield the distances between fluorescent dyes, which are commonly used for labelling proteins, DNA, RNA, etc. If multiple FRET experiments are done, it is possible to build a 3D model of the labelled positions. FRETsg finds all positions which obey the measured FRET distances and writes the model to a PDB-type file, which can be easily overlayed with other PDB structures.
FRETsg minimizes the following expression using a steepest descent method and starting from a random configuration of points:
where N is the number of points, whose positions should be found by this minimization; D is the number of FRET distances; Kij is the force constant, which is given by the width of the distance distribution; ri is the position of point i, rij0 is the measured FRET distance between point i and j.
This minimization procedure is done several times, starting from random configurations, to find all configurations that obey the given FRET distances.
After the minimization FRETsg performs a Monte-Carlo simulation in the energy landscape given by the expression above.
Publication
Installation
License Agreement
Unix/Linux
To install FRETsg, move the tar-file to your favourite directory and type:
- for Linux: tar xvzf fretsg-1.0.tar.gz
- for Unix: gunzip fretsg-1.0.tar.gz gtar -xvf fretsg-1.0.tar
The source is just one ANSI-C file without any system specific stuff. It should compile with: cc -O2 -o fretsg fretsg.c -lm
Windows
Just unzip the zip-file into your favourite directory, this will unpack the executable fretsg.exe.