How to Perform Constant pH Simulations in GROMACS
Here we provide a version of Gromacs 3.3.1 and of Gromacs 3.3.3 for performing constant pH MD as described in our publications Donnini et al. (2011) and Dobrev et al. (2017). The next version of the constant pH MD, which should be easier to use and faster, in Gromacs 4.5 will be available in the future. The constant pH MD in Gromacs 3.3.1 and Gromacs 3.3.3 can be used ONLY with PME, Cut-off or Reaction-Field types for electrostatics and the Cut-off type for van der Waals interactions. At the moment ONLY a change of the nonbonded terms (charges and van der Waals) and masses upon protonotation/deprotonation is possible. Note also that a change in the van der Waals (i.e., atom types) is correctly evaluated ONLY for chemically uncoupled titrating sites (see below two states model). If you want to use other electrostatics and/or van der Waals types please contact Plamen Dobrev. Note that the TIP4P water model is not compatible with the current version of constant pH MD in Gromacs 3.3.1 and 3.3.3.
We also provide a "1lambdapernode" version for constant pH MD in Gromacs. The "1lambdapernode" is faster than the non-1lambdapernode version when many ionizable groups are included. You can read more about this version below.
For running constant pH MD with neutral-charge downlowad the "_neutral" version of the code. Here, we impose a constraint on the total number of protons of the system. For details about the method, please refer to Donnini et al. (2016).
Compilation of the code is the same as for the standard version of Gromacs.
Current status of the implementation at the interal department Wiki.
Setting up a constant pH simulation
Before starting a constant pH MD, you need to decide which are the ionizable groups you want to consider in the simulation (e.g., all ionizable amino acids of your protein). Note that for N ionizable groups the constant pH MD with Gromacs 3.3.1 and Gromacs 3.3.3 is about N time slower than a standard free energy simulation of the deprotonation of one such groups. This is different for the "1lambdapernode", which is explained further below. For each type of ionizable group you need to define a reference system (generally the free amino acid in solution). For starting the constant pH MD a) the experimentally measured and b) the calculated deprotonation free energy of the reference system are required (reference simulation - see below).
In the most simple case an ionizable group is described by TWO states (i.e. H=protonated and /=deprotonated) and a titration coordinate lambda (l). In this case you will need the topology of the protonated and deprotonated groups. For example, for histidine a two state model description means that one of the nitrogens of the side chain, always the same during the simulation, can be protonated or deprotonated during the constant pH simulation.
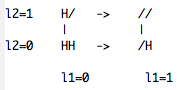
To simulate deprotonation/protonation of both nitrogens on the side chain of histidine, we use a FOUR state model where histidine exists in the four different protonation states (HH,H/,/H and //). The reason for having these four states is that the two nitrogens on histidine's side chain are chemically coupled, i.e., deprotonation/protonation of one nitrogen changes the chemical attributes (and therefore the topology) of the titratable site on the other nitrogen. In this case the topology of each state of histidine and two titration coordinates lambda (one for each titrable site) are required.The two and four states models are described in the reference [1]. Note that in the constant pH MD in the Gromacs 3.3.1 and Gromacs 3.3.3 versions a change in the bonded terms is implemented ONLY for the two states model. To help building the topology and setting up the reference simulations (see below) for a four states model build a scheme of the four states, which are connected by two lambda coordinates lambda1 (l1) and lambda2 (l2) as shown in the first scheme.
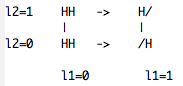
If you do not wish to include one of the four states, for example for histidine the fully deprotonated histidine (//), you can use the four states model where the four states are now defined as HH,HH,H/ and /H. In this case one of the titration coordinates (l2) act as a switching coordinate between the H/ and /H states. The scheme to follow in this case is shown in the second scheme.
During deprotonation the charge of the system changes. This can lead to artefacts in your simulation, in particular for protein/membrane simulations with PME. Please refer to publications by Hummer et al. (1996), Hünenberger et al. (1999), or Börjesson et al. (2001) for examples. You can decide to couple an ion/hydronium to the ionizable group, so that the overall charge stays constant. In this case you should avoid interactions between the ion/hydronium and the amino acid. Generally beyond 1.5 nm distance between amino acid and ion/hydronium (at 0.150 M salt concentration) there are no more interactions. For more details about this model, please ask Plamen Dobrev.
Reference free energy simulation
To calculate the deprotonation free energy of the reference system we run a free energy simulation. For the ionizable groups of a protein the reference system is generally the single (methyl-capped) amino acid in bulk water. If the ionizable group is coupled to an ion/hydronium, the A and B states of the atoms of the ion/hydronium are also required. Generally a thermodynamic integration (TI) is performed between the protonated and deprotonated states. To improve accuracy, perform the TI in the lambda range -0.05 to 1.05, instead of 0 to 1. All conditions of the reference free energy simulation (i.e., A and B states of the ionizable group, force field, mdp file options, ionic strength) must be the same in the reference and in the constant pH simulation. If the box shape and/or size are different in the reference and constant pH simulations, make sure this does not introduce different contributions to your free energy. Please refer to publications by Hummer et al. (1996), Hünenberger et al. (1999), or Börjesson et al. (2001) for examples. Plamen Dobrev has checked the effect of increasing box size on the free energy for deprotonating a methyl-capped aspartic acid in 0.150 M salt concentration using PME. The changes are small (about 0.5 kJ/mol) with increasing box size from 3 nm to 9 nm.
Once the deprotonation free energy is calculated, we perform a linear or third order polynomial fit to the dV/dlambda profile obtained from the TI. The coefficients of this polynomial are what you need for the constant pH simulation.
For ionizable groups of a protein described with a FOUR state model more free energy simulations are required. This is because the deprotonation free energy along one titration coordinate lambda depends on the value of the second coordinate lambda. For example for histidine, the calculated reference deprotonation free enery for the ND1 titration site is obtained from 11 TIs. In the first TI the A and B states correspond to ND1 protonated and deprotonated, respectively, and NE2 always protonated, and in the last TI to ND1 protonated and deprotonated, respectively, and NE2 always deprotonated. A polynomial is fitted to each of the 11 dV/dlambda profiles. The 0th, 1st, 2nd and 3rd order coefficients of these polynomials are in turn fitted to other 4 polynomials. One obtains therefore 16 coefficients which are needed for the constant pH simulation. Similarly is done for the reference deprotonation free energy of the NE2 titration site.
The reference free energy simulations can now be run using version 1.4 of the constant pH MD (download Const_pH_v1.4_Gromacs 3.3.3). By setting the option lambda_dynamics_ftype=5 in file.mdp, the constant pH MD runs keeping the values of lambda equal to the initial_lambda values set in the input file (see below). The dV/dlambda for each lambda groups is printed in the standard output file. For the reference free energy simulations of a four state model titratable group this procedure is advised, with respect to using the standard free energy code. The reason for this is that setting the free energy simulations by interpolating manually charges along one lambda in the topology, does not results in the same potential energy surface at intermediate lambda values, as by interpolating the potentials (as in done during the constant pH MD).
Note that to obtain energy conservation for a four state model, it is advised to fit a function to the DG potentials, rather than to the dV/dlambda. For more details, please contact Plamen Dobrev.
Constant pH MD simulation
The topology and mdp files for running a constant pH MD with Gromacs 3.3.1 and Gromacs 3.3.3 are described below. You will also need to have in your running directory three additional files (lambda_groups.dat, pka_data.dat, and ph.dat) when using a two states description of the ionizable group, or more for a four states description. Two input files (lambda_groups.dat, pka_data.dat) differ between the Gromacs 3.3.1 and Gromacs 3.3.3 constant pH MD versions. These are described below.
INPUT FILES for constant pH MD Gromacs 3.3.1
Here a list of the input files for the TWO and FOUR states model. You must use the same format as shown in the example file (including punctuation and order in which the options are listed) for lambda_groups.dat, pka_data.dat, ph.dat, param_xxxx.dat and xxxB.top files.
The Two States Model
topology file
The topology is the same as for a standard free energy calculations with Gromacs. State A and B have to be defined, e.g., A is protonated and B deprotonated, for each of the ionizable aminoacids which you want to include. The A and B states of one ionizable group are defined in the same way as in the reference free energy simulation of the reference system of that group. If the ionizable group is coupled to an ion/hydronium, the A and B states of the atoms of the ion/hydronium are also required. For histidine or any other aminoacid (e.g., glutamic acid where both oxygens are able to protonate) where there are more then two states see description further on for four states model.
md.mdp file
There are five additional fields in the free energy part of the mdp file, and one additional field for setting the seed for the andersen thermostatting. Note that the free energy option has to be "yes".

lambda_dynamics: yes (switches on the lambda-dynamics)
lambda_dynamics_ftype: 4
nu_T_lambda: coupling time of the lambda to the andersen temperature
bath (a value of nu_T_lambda = 6 means: Every sixth timestep).
T_lambda: the temperature (in Kelvin) of lambda.
m_lambda: the mass of lambda (in u).
andersen_seed: the start seed of the random number generator used for the temperature coupling of lambda to the andersen thermostat. 0 means that at every run a random number is used as start seed.
lambda_groups.dat file.
Here all the ionizable groups are listed.

name: MUST be a four letter name! It can be different from the residue name in the topology file, but has to be the name as in the example pka_data.dat file.
residue_number: number of the residue.
initial_lambda: between 0 and 1.
number_of_atoms: number of atoms of the ionizable group which are perturbed -i.e., for which an A and B state is defined-. Remember to include the atoms of the ion/hydronium if there is one coupled to the ionizable group.
list of atom number: this is a list of the atom number of the atoms of the ionizable group according to the coordinate file (e.g., file.gro). Again, remember to include the atoms of the ion/hydronium if there is one coupled to the ionizable group.
pka_data.dat file.
In this file the parameters of each titratable group are listed. Note that if you have, for example, 3 glutamic acids in the protein (and in the lambda_groups.dat file), you only need one entry for glutamic acid in the pka_data.dat file.
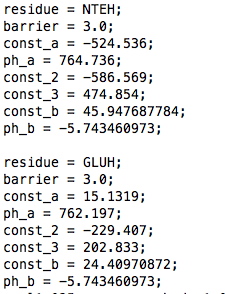
residue: must correspond to the name in the lambda_groups.dat file.
barrier: this is the height (in kJ/mol) of a parabolic barrier between the 0 and 1 states of lambda (if you wish a different barrier shape read about the constant pH Gromacs 3.3.3 version).
const_a, ph_a, const_2, const_3: these correspond to the coefficients (up to the third oder, with const_a order 0, ph_a order 1, const_2 order 2 and const_3 order 3) of a polynomial which is fitted to the dV/dlambda of the free energy simulation of the reference system (for a linear fit, const_2=0, const_3=0). To improve accuracy, it is advisable to calculate this dV/dlambda in the range -0.05 to 1.05, instead of 0 to 1.
const_b: this is ln(10)*R*T*pKa_ref, with R the gas constant (in kJ/(mol Kelvin)), T the temperature (in Kelvin) and pKa_ref the experimental pKa of the reference system.
ph_b: this is -(ln(10)*R*T), with R the gas constant (in kJ/(mol Kelvin)) and T the temperature (in Kelvin).
ph.dat file.
Here the pH is listed. For example
7.0;
The Four States Model
topology files (topol.top, HISB.top)
For the four states model the topologies of four states are required. In the topology file topol.top only two states are defined. For example, for histidine, the A and B state in the topol.top file describe the fully protonated (HH) and the singly deprotonated (H/, e.g., depronated on ND1) histidine. The topology for the other two states (/H and //) has to be written in a separate file which needs to be in the running directory. This file must be named HISB.top and contains charges and atom types for the singly deprotonated histidine (on NE2) and for the fully deprotonated histidine, in the format of the example file provided here. If you wish to describe more titratable sites, other than histidine, with the four state model, contact Plamen Dobrev.
md.mdp
This is the same as already described for the two states model.
lambda_goups.dat

For the example of histidine, the two titratable sites on the side chain must be called HISA (e.g., ND1 site) and HISB (e.g., NE2 site), and they will consist of the same atoms, i.e., all the perturbed atoms of histidine. See the example file shown here. HISA MUST be listed before HISB.
pka_data.dat
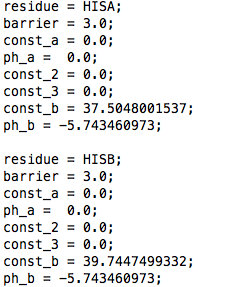
There must be an entry for HISA and one for HISB in the pka_data.dat file (see example here). The const_b field of this file contains the measured reference microscopic pKa for that titratable site - see for example reference Tanokura (1983). Note that we use 14.1 as value for the second reference microscopic pKas of ND1 and NE2. If you wish to change this value, contact Plamen Dobrev. The const_a, ph_a, const_2, const_3 fields are not read from the pka_data.dat file for the four state model. Instead, two additional files must be in the running directory. These are called param_HISA.dat and param_HISB.dat and are explained below.
param_HISA.dat, param_HISB.dat
The param_HISA.dat and param_HISB.dat files contain each 16 coefficients which are obtained with the fitting procedure described above in "Reference free energy simulation" for each titration site of histidine (i.e., nitrogen ND1 and nitrogen NE2). For example, for HISA (e.g., nitrogen ND1) which is described by the titration coordinate lambda1, dV/dlambda1 is fitted to a third oder polynomial
dV/dlambda1 = a0 + a1*lambda1 + a2*lambda1^2 + a3*lambda1^3
with
a0 = a00 + a01*lambda2 + a02*lambda2^2 + a03*lambda2^3
a1 = a10 + a11*lambda2 + a12*lambda2^2 + a13*lambda2^3
a2 = a20 + a21*lambda2 + a22*lambda2^2 + a23*lambda2^3
a3 = a30 + a31*lambda2 + a32*lambda2^2 + a33*lambda2^3
with lambda2 the titration coordinate for the HISB titratable site on the other nitrogen of the side chain of histidine.
The coefficients are listed in the param_HISA.dat and param_HISB.dat files in the following order
a00 a01 a02 a03
a10 a11 a12 a13
a20 a21 a22 a23
a30 a31 a32 a33
ph.dat
Same as for the two states model.
The Three States Model
As mentioned above, the four states model can also be used if you do not wish to include all four protonation states for two chemically coupled titrating sites, such as the two nitrogens on the side chain of histidine. For example when describing glutamic acid or aspartic acid where both oxygens of the carboxylic group can deprotonate, one can exclude the fully protonated state which will practically not occure. The residue names to identify this description are four letter names which end with 2A and 2B: xx2A and xx2B, i.e., GL2A and GL2B for glutamic acid, AS2A and AS2B for aspartic acid, HI2A and HI2B for histidine. These names do not need to correspond to the names in the topol.top and coordinate file but only in the additional files (lambda_groups.dat, pka_data.dat, param_xx2A.dat, param_xx2B.dat and xx2B.top). If you wish to include more residues which are described by a three state model contact Plamen Dobrev.
INPUT FILES for constant pH MD Gromacs 3.3.3
The input files are all the same as described for constant pH MD with Gromacs 3.3.1, except for lambda_groups.dat and pka_data.dat. In lambda_groups.dat there are four additional fields. The first three concern the height (barrier_l2_0, barrier_l2_1) and shape (points) of the barrier potential (or biasing potential) between the protonated and deprotonated states of lambda. In particular, in the constant pH MD Gromacs 3.3.3 version, the barrier height for the four/three states model is given by
barrier of lamba = (1-lambda2)*barrier_l2_0 + lambda2*barrier_l2_1
If you are using a two states model or you don't want to make use of this, just set barrier_l2_0 and barrier_l2_1 to the same value.NOTE: at the moment this barrier option is still unser test. It is advised to use barrier_l2_0=barrier_l2_1.
The field "points" is used to build barrier potentials of certain shapes. Points indicates the number of points (maximum 7) which are used to build a spline function. By setting points to zero, a parabolic barrier is used. At the moment the points have to be changed in the program code. If you want to use this field contact Plamen Dobrev
lambda_groups.dat
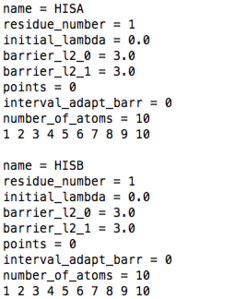
name: as for constant pH MD Gromacs 3.3.1
residue_number: as for constant pH MD Gromacs 3.3.1
initial_lambda: as for constant pH MD Gromacs 3.3.1
barrier_l2_0: barrier height (in kJ/mol) of a barrier potential between the 0 and 1 states of lambda when lambda 2 is zero
barrier_l2_1: barrier height (in kJ/mol) of a barrier potential between the 0 and 1 states of lambda when lambda 2 is one
points: set the number of points used to build a spline function that replaces the parabolic barrier. When points is equal zero a parabolic barrier is used. If you want to use points other than zero contact Plamen Dobrev.
interval_adapt_barr: this must be zero
number_of_atoms: as for constant pH MD Gromacs 3.3.1
list of atom number: as for constant pH MD Gromacs 3.3.1
pka_data.dat
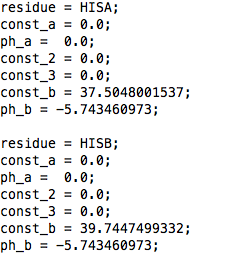
Same as for constant pH MD Gromacs 3.3.1, BUT the field "barrier" is absent.
OUTPUT FILES
The output files are the same for constant pH MD in Gromacs 3.3.1 and Gromacs 3.3.3. The output file is called l_dynamics_group_x.dat with x the number of the group in the lamda_groups.dat file (i.e., the first group listed in lambda_groups.dat gets number 1). Here the time and lambda are printed every 500 steps (and for the 3.3.3 also an additional column which is always zero). The constant pH MD also prints the dV/dlamba and the lamba of each group in the standard output file every step (can get quite large...). In the beginning of the standard output some information about reading of the input files is also printed. This information is however printed by each processor. Therefore, if you want to check if input files are read correctly run on one processor a one step only constant pH simulation.
Constant pH MD "1lambdapernode" with Gromacs 3.3.1 and Gromacs 3.3.3

The "1lambdapernode" version calculates the forces acting on each ionizable group on a different processor (replica). For N ionizable groups and p processors there will be N/p groups per processor (you can check this in the standard output). Therefore, if you have e.g. 30 groups, you will be better off by using 30 processors. Note however that if you have e.g. 4 ionizable groups you will be faster with the non-1lambdapernode (if you run with more than 4 processors) as the speed of the 1lambdapernode version will not increase with more than 4 processors.
In the graph, we compare the performances of the non-1lambdapernode and 1lambdapernode versions for a system of 32 ionizable groups (448 perturbed atoms in total; system size=23,000 atoms) using PME for long range electrostatics. Due to the different scaling, the performances will be different when using a different scheme for electrostatics.
The input and output files are identical to the ones already explained above. To use this version you have to use the replica exchange option of Gromacs 3.3.1 and Gromacs 3.3.3 and set replex to zero
mdrun -multi -replex 0
Publications and References










