Conformational Flexibility of Prions
Helmut Grubmüller, Martin Billeter, Kurt Wüthrich
Support: EMBO (short term fellowship for H.G.)
The conformational plasticity of mouse proin protein fragment PrPc (cellular isoform), the structure of which has been solved by NMR [1], has been characterized by molecular dynamics simulations. In the simulations, the method of 'conformational flooding' [2] has been used to destabilize the cellular isoform and thereby to introduce conformational transitions. An opening motion of helix 1 and helix 2 was observed as a first step which increases solvent exposure of helix 3. That opening motion is triggered by the rupture of hydrogen bolds which bind the beta sheet to helix 1. A similar, but spontaneous opening motion was observed in a simulation of the T183A mutant, one of the inheritable Creutzfeld-Jakob disease (CJD) sites. As a subsequent conformational change, our simulations suggest a klink of helix 2, accompanied by extension of the initially short beta sheet towards helix 1 in agreement with a recently proposed model. Five 'hot spots' of the induced conformational transitions have been determined as regions where large sliding motions occurr; three of these correlate perfectly with the inheritable CJD sites Val 61, Asp 59, and Tyr 99. We therefore suggest the described conformational transitions as a model for the initial steps of the pathogenic transition towards the scrapie isoform PrPSc.
Introduction
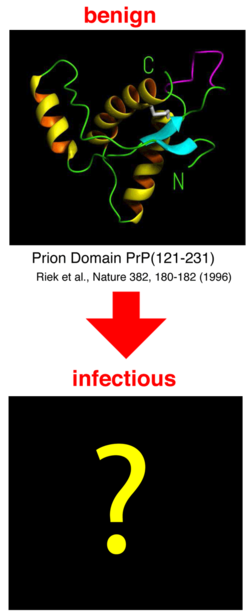
According to the 'protein only hypothesis', several neurodegenerative diseases - e.g., BSE (bovine spongiform encephalopathy), Creutzfeldt-Jakob disease in humans, and scrapie in sheep - is triggered by a structurally (i.e., not chemically) modified version of prion protein. A structural transition of the non-pathogenic conformation, PrPC, to the pathogenic conformation, PrPSc, seems to be the ultimate molecular cause of the diseases. Therefore, an understanding of the microscopic transition mechanism as well as knowledge of the PrPC and particularly of the PrPSc structure is essential and can contribute to the development of medications.
Here we describe the results of molecular dynamics simulations which aimed at the identification of 'soft' deformation motions of the known PrPCstructure, and which would provide a model for the initial steps of the pathogenic structural transition.

Methods: Simulation System
Starting from the X-ray structure (PDB entry 1ag2) we placed the proin protein within a sphere of 8691 TIP3 water molecules with 80 Å diameter. All simulations were carried out using the parallel MD program EGO which employs the CHARMM force field and allows efficient computation of electrostatic interactions. The system comprised 27144 atoms.
After minimization, the system was equilibrated by performing free dynamics simulations for 360 ps. The resulting system was used as starting point of the subsequent conformational flooding simulations. In total, the system was simulated for 7.5 ns.
Methods: Flooding
A series of flooding simulations was carried out to probe which conformational transitions of the protein can be induced most easiest. The basic assumption is that these are similar to the first steps of spontaneous or catalyzed pathogenic conformational transitions.
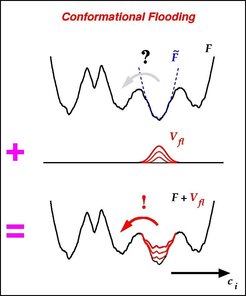
The flooding potential is derived from a harmonic model F~; (dashed line) of F in the vicinity of the initial NMR conformation (local minimum). F~ is computed from an ensemble of structures extracted from an MD simulation.
Inclusion of the localized flooding potential Vfl into the molecular force field (Figure, bottom) raises the free energy minimum that defines the initial conformation substrate without significantly affecting adjactent energy barriers. Accordingly, the initial configuration is destabilized and conformational transitions out of the initial susbstrate are accelerated such that the (typically slow) transition rate is shifted into the fast MD simulation regime of several 100 picoseconds and can be studies within such 'flooding simulations'.
Within each of the flooding simulations the (high dimensional) free energy landscape F to the protein (Figure, top) was artificially perturbed by including a 'flooding potential' Vfl (Figure, mid) of adjustable strenght into the molecular force field.
Results: Control
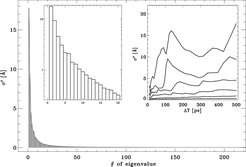
Few collective degrees of freedom dominate the atomic fluctuation in the prion protein. A fluctuation analysis has been carried out for the structure ensemble that was used to construct the flooding potential. The obtained eigenvalue spectrum indicates the amount of atomic motion described by the corresponding degree of freedom.
To check if the used ensemble is large enough to describe the initial conformational substrate accurately, the eigenvalue spectrum has been determined on the basis of ensembles of varying size. The right inset shows (from top to bottom) the values of eigenvalues #1, #2, #3, #5, #10 and #20, respectively, as a function of the length of the MD simulations from which the respective ensemble was extracted. For construction of the flooding potential, an ensemble of size ΔT= 300 ps was used.
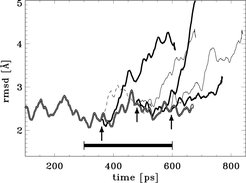
Smoothed rmsd deviations from NMR structure during equilibration (double line) and during flooding (six single lines). The flooding simulations started at the instances indicated by the three arrows.; at each instance two flooding simulations were carried out, with flooding strength of 50 kbT (thin lines) and 100 kbT (bold lines), respectively. The abrupt increase of rmsd deviations reflects conformational transitions induced by the respective flooding potentials.
The dashed line indicates a simulation, during which no conformational transition was observed. The ensemble that was used to construct the flooding potential was extracted from the trajectory piece denoted by the horizontal bar.
Results: Structural Transitions
Models for two subsequent structural transitions of prion protein were condensed from a total of 15 flooding simulations that have been carried out:
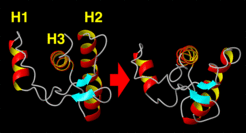
Step 1: Opening Motion and Helix Exposure
All of the flooding simulations showed an initial opening motion of helix 1 (H1) and helix 2 (H2), thereby increasing exposure of helix 3 (H3) to the surrounding water solvent.
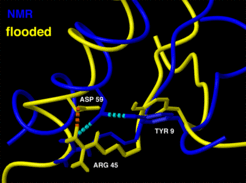
This conformational transition is apparently triggered by rupture of two hydrogen bonds, Tyr9 - Asp 59 and Arg-45-Asp59, respectively, which in the NMR structure bind to the beta sheet to helix 1.
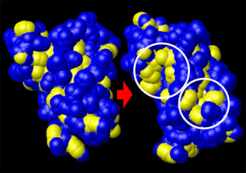
Exposure of hydrophobic groups . Left: equilibrated NMR-structure, right: structure after transition. shown is the solvent accessible surface of the protein; CHn-groups belonging to hydrophobic residues are highlighted yellow. Circles indicate regions of significant exposure. The hydrophobic surface increased from 1759 Å2 to 2121 Å2. Assuming a free energy cost of 0.021 kcal/mol/Å2 to create hydrophobic surface, the structural transition would decrease solubility by 7.6 kcal/mol.
Step 2: Helix Kink and Beta Sheet Extension
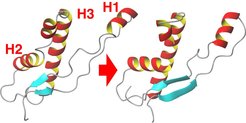
In some of the flooding simulations a second structural transition occurred, which involved (a) breakage of helix 1 and (b) extension of the (blue) beta sheet towards helix 1. This finding supports a model for the pathogenic PcPC → PrPSc conversion that assumes the beta sheet to be a nucleation center for the transformation of helix 1 into a beta sheet.
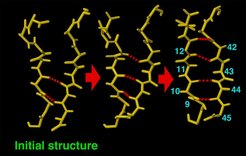
In the course of the second major conformational transition induced by the flooding potential, two additional beta sheet hydrogen bonds (red) built up sequentially. only the residues in the vicinity of the beta sheet are shown.
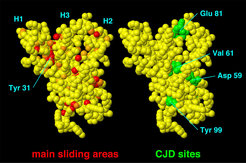
'Hot spots' of the structural transitions extracted from our flooding simulations (colored red, left side) correlate with three mutations have been identified in cases of inherited CJD (green, right side). These are Asp 59, Val 61, and Tyr 99. From the simulations, the 'hot spots' have been derived as main sliding regions, for which van der Waals contacts between close atoms change most often. One 'hot spot' (Tyr 31) appears to be unrelated to known CJD sites. Unfolding experiments of Tyr 31 mutants is therefore suggested, which are expected to demonstrate reduced stability.
Summary and Conclusions
A stwo-step model for the initial structural transitions of the pathogenic PcPC → PrPSc conversion has been proposed. The model involves mild solvent exposure of helix 3, a subsequent kink of helix 2, and extension of the beta sheet. It resembles a proposed destabilizing effect of a CJD mutant (T183A) for the wild type. In our model three out of four known CJD sites are identified as 'hot spots', i.e., centers of large sliding motions; accordingly, we explain the effect of the mutants in terms of a decreased resistance against these sliding motions.










