Unbinding Simulations of the AN02-Antibody-DNP-Hapten Complex
Berthold Heymann and Helmut Grubmüller
Collaborators: Wolfgang Dettmann, Angelika Wehle, and Hermann Gaub (LMU Munich).
Support: DFG (SFB 143/C2)
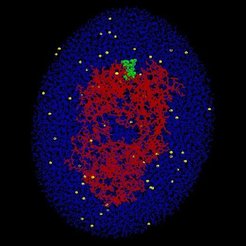
The unbinding of the AN02-antibody-(dinitrophenyl [DNP]-hapten) complex was studied by molecular dynamics (MD) simulations. In particular, we focused at the antigen binding Fab-fragment of the AN02-antibody, where the spin-labeled DNP-hapten is bound to. In order to account for solvent effects, the AN02-hapten complex (red/green) was "solved" in a water droplet (blue) including ions (yellow) in physiological concentration. The whole system comprises about 45,000 atoms. In close resemblance to atomic force microscopy (AFM) unbinding experiments, the oxygen atom (O2) of the hapten was attached to a harmonic potential (symbolized by a spring), which models the cantilever of an atomic force microscope. In the course of the simulations, the spring was moved towards the right (arrow) with constant pulling velocity vcant.
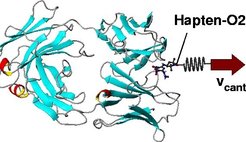
Thus, an increasing pulling force was exerted onto the O2-atom and thereby onto the whole hapten molecule, and unbinding motions where induced. To prevent that the whole complex was pulled towards the right, the center of mass of the AN02-antibody-Fab- fragment was kept in place by a stiff harmonic potential. Internal motions of the antibody as a reaction of the unbinding process remained unaffected.

In the course of the simulations the pulling force was recorded as a function of the equilibrium position of the spring (the cantilever position) yielding a force profile for each unbinding simulation. Cantilever position 0 Å corresponds to the bound state, 15 Å to the unbound state. The maximum of this force profile was attributed to the unbinding force of the complex, which is to be compared to experimental data (not yet available). To leave the binding pocket the ligand has to overcome several force barriers. MD simulations and AFM experiments take place at significantly different time scales: whereas MD simulations cover at maximum tens of nanoseconds, AFM experiments are carried out at the millisecond time scale and slower. Accordingly, in the MD simulations the hapten molecule was pulled out of the binding pocket with much larger pulling velocities.
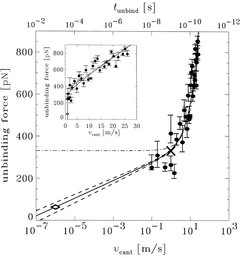
To study the dependence of the unbinding force on pulling velocity and to compare the calculated with measured unbinding forces, a series of unbinding simulations with pulling velocities ranging from 0.1 m/s to 30 m/s were performed. Especially from the inset, with shows the unbinding force as a function of pulling velocity in a linear plot, it is visible that for pulling velocities above 3 m/s a linear dependence is dominant. This can be interpreted as Stokes' friction. For pulling velocities below 3 m/s, however, a logarithmic dependence dominates, as can be seen from the logarithmic plot. This can be explained as consequence of activated processes due to thermal fluctuations, which decrease the free energy activation barrier and become relevant at low pulling velocities.
Using a model, which describes friction and activated processes, and which makes use of the experimentally determined spontaneous dissociation rate of the AN02-hapten complex, the calculated unbinding force can be interpolated to the experimental millisecond time scale (solid line). From this interpolation we expect an unbinding force of 60 +/- 30 pN for AN02-hapten unbinding experiments, which are being carried out in Hermann Gaubs lab in Munich.
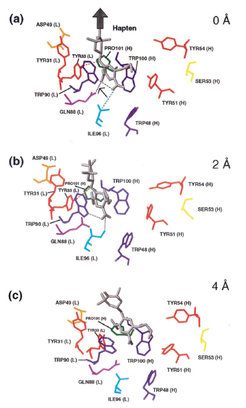
Our MD simulations allow detailed insight into the processes that occur during the unbinding process at the atomic level.
In the bound state (top) the hapten molecule (grey) is located within a tryptophane sandwich, which cause strong van der Waals contacts between the hapten DNP-ring and the aromatic rings of the tryptophane residues. This configuration is stabilized by a couple of hydrogen bonds (dotted lines). After the cantilever was moved 2 Å in pulling direction (big arrow), the first strong hydrogen bond (between GLN88 (L) and one of the nitro groups of the DNP-ring) was broken and the interaction between the whole hapten molecule and the binding pocket lowered. However, as visible from the force profile, the maximum force barrier (the unbinding force) has not yet been crossed. Pulling the cantilever another 2 Å in pulling direction (bottom) caused the breakage of the remaining hydrogen bonds and the release of the hapten DNP-ring out of the tryptophane sandwich. In this state, the maximum force barrier has just been crossed.
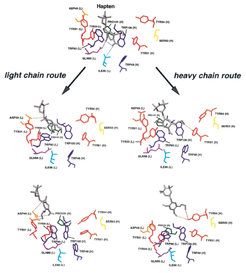
Our unbinding simulations revealed that the hapten molecule can move along the light chain (snapshot on the left) as well as along the heavy chain (snapshots on the right) out of the binding pocket. Which route was chosen in a particular unbinding process was hihgly dependent on what happened first: the release of the hapten molecule out of the tryptophane sandwich or the breakage of the hydrogen bond betwenn TYR33 (L) and the nitrogen atom in the middle of the hapten. When the hydrogen bond remained intact, after the DNP-ring had moved out of the tryptophane sandwich, the whole hapten molecule showed the tendency to move towards the light chain, otherwise the route along the heavy chain was favorable. Thus, the hydrogen bond to TYR33 (L) can be interpreted as a switch, which strongly affected the unbinding route. The snapshots show that the interactions between the hapten molecule and the binding pocket residues during the unbinding process (dotted lines) are significantly different for the light chain route and the heavy chain route.
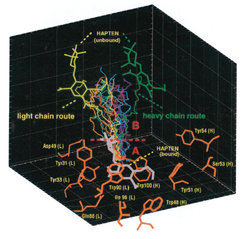
Our simulations revealed that there are not only the two extreme routes out of the binding pocket (residues shown in orange), but a whole spectrum of unbinding pathways (coloured lines); for the light chain route and the heavy chain route, the unbound hapten molecule is shown explicitly (yellow and green, respectively). This is what we call structural heterogeneity of the unbinding pathways. Structural heterogeneity means that there are a couple of possibilities for the hapten to move out of the binding pocket, and, expressed in a more abstract way, that a comparatively large volume in the configurational space can be accessed. This implies, in turn, that entropy significantly contributes to the bond. This contribution we estimated to 4 kcal/mol (not shown) compared to an experimentally determined free energy difference of about 16 kcal/mol for the AN02-DNP-hapten complex.
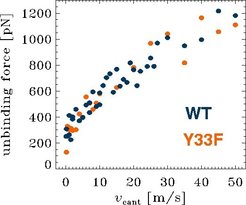
To study the role of individual residues of the binding pocket to the unbinding process, we carried out a series of unbinding simulations for a mutant, where the TYR33 (L) was replaced by a PHE. In contrast to TYR in the wildtype, PHE does not form a hydrogen bond to hapten. Accordingly, for the Y33F mutant lowered unbinding forces were expected. However, as visible from the plot, the Y33F mutant unbinding forces (orange) do not signigficantly differ from the wildtype unbinding forces (blue). What is the reason for this surprising finding? Why do the unbinding forces remain more or less unchanged, although one stong hydrogen bond was "switched off"?
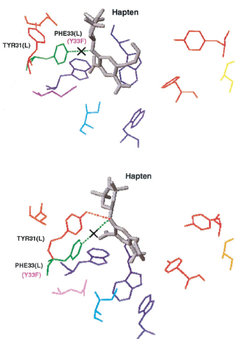
The snapshot to the right gives the answer. In the bound state (top) of the Y33F mutant there is indeed one hydrogen bond less than in the wildtype complex (dotted line crossed out). However, when the hapten was partially pulled out of the tryptophane sandwich (bottom), the neighboured TYR31 (L) moved towards the hapten and formed a hydrogen bond to the nitrogen atom in the middle of the hapten. Thus, the TYR31(L) replaced the lacking TYR33(L), and there was more or less the same net interaction strength between hapten and the AN02 binding pocket. Accordingly, the unbinding forces have not changed significantly.








