U4 snRNA Kink-Turn
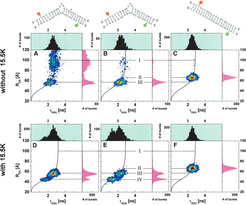
The kink-turn (k-turn), a RNA structural motif found in the spliceosome and the ribosome, serves as a specific protein recognition element and as a structural building block. Whereas the structure of the spliceosomal U4 snRNA k-turn/15.5K complex is known from a crystal structure, it is unclear whether the k-turn also exists in this folded conformation in the free U4 snRNA. Thus, we investigated the U4 snRNA k-turn by single-molecule FRET measurements in the absence and presence of the 15.5K protein and its dependence on the Na+ and Mg2+ ion concentration.
We show that the unfolded U4 snRNA k-turn introduces a kink of 85–15 in an RNA double helix. While Na+ and Mg2+ ions induce this more open conformation of the k-turn, binding of the 15.5K protein was found to induce the tightly kinked conformation in the RNA that increases the kink to 52–15. By comparison of the measured Förster Resonance Energy Transfer (FRET) distances with a computer-modeled structure, we show that this strong kink is due to the kturn motif adopting its folded conformation. Thus, in the free U4 snRNA, the k-turn exists only in an unfolded conformation, and its folding is induced by binding of the 15.5K protein.
