Stiffness of the Juxtamembrane Region of the t-SNARE Syntaxin 1A
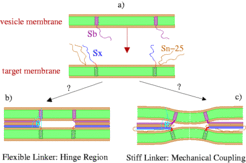
For exocytosis and for the selective transport of macromolecules between the various organelles of eukaryotic cells the merging of a transport vesicle membrane with a target membrane is an essential step.
According to the SNARE hypothesis, this process is mediated through the assembly of vesicle (v) and target (t) SNARE proteins [1] (Fig. 1 a). The cytosolic core of the SNARE complex involved in synaptic transmission has been resolved by X-ray crystallography and consists of the v-SNARE synaptobrevin-II (magenta) and the t-SNAREs syntaxin 1A (blue) and SNAP-25 (brown) which form a four-stranded parallel coiled coil [2] (Fig. 1 b,c). A 5-residue basic linker of unknown structure (red) connects the syntaxin 1A H3 helix (blue) with its c-terminal membrane anchor (green) which can be assumed to have alpha-helical conformation. For the initial step of membrane fusion, two scenarios are currently considered and discussed controversally [3,4]: (b) The linker is very flexible and represents a hinge region between the two adjacent syntaxin domains. (c) The linker is stiff enough to provide a mechanical coupling of the two syntaxin domains; as the SNAREs assemble, the target membrane is bent towards the vesicle membrane.
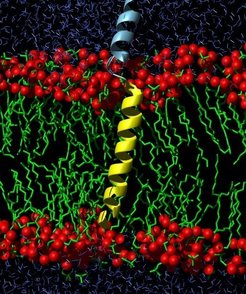
The present simulation study aims to contribute to this discussion by computing the stiffness of the linker. The stiffness of the linker is studied by molecular dynamics (MD) simulations including an 11-residue c-terminal part of the syntaxin 1A core helix (X-ray helix, turquoise).
To stabilize the known secondary structure, internal restraints were imposed on the X-ray helix. Two linker residues adjacent to the X-ray helix were found to spontanously fold from loop into alpha-helical conformation and therefore for the presented simulations were also kept in alpha-helical conformation by internal restraints. The three remaining residues were modeled in random loop conformation.
- its 5-residue basic linker (grey loop) and its 23-residue alpha-helical transmembrane domain (yellow).
- The embedding lipid bilayer comprising 116 lipid molecules (green, the oxigen atoms of the polar region are shown as red spheres).
Three different lipid species were incorporated:
- Palmitate-Oleate(PO)-Phosphatidylcholine (PC): zwitterionic
- PO-Phosphatidylethanolamine (PE): zwitterionic
- PO-Phosphatidylserine (PS): acidic
These lipid species were combined to obtain two different lipid compositions:
- 70 POPC and 46 POPE (yielding a neutral bilayer)
- 70 POPC, 34 POPE, and 12 POPS (yielding a negatively charged bilayer).
The lipid bilayers were obtained by modifications of an equilibrated bilayer of POPC molecules.
- An explicit solvent environment (water molecules shown in blue). The solvent environment comprised 4720 water molecules and (dependent on the lipid composition) 6 chloride or 6 sodium ions such as to counterbalance the netto charge of the system.
All molecular dynamics simulations are carried out using the GROMACS simulation package [5]. The systems are simulated at 300K and a pressure of 1 bar with periodic boundary conditions. The free energy for peptide bending is estimated from the equilibrium fluctuations. Preliminary results show the linker to be surprisingly stiff: To bend the linker by 20 degrees requires approx. 2 kcal/mol. Therefore our simulations so far support the mechanical coupling scenario.

