A Highly Dynamic Folding Intermediate of Ubiquitin in a Force Clamp

Collaboration: Julio Fernandez (Columbia University, NY, USA)
Financial Support: Volkswagen Foundation and Boehringer Ingelheim Fonds.
Proteins enable living organisms to move, metabolize and sense, in short, to function. To fulfill such diverse tasks, proteins take up a specific fold, their native structure. The factors driving the complex folding process are not yet fully understood. Conventional studies subject proteins to denaturing conditions such as elevated temperatures, acidic pH, or urea as denaturant and observe changes during refolding, such as fluorescence quenching, or amide bond vibrations.
Recently, single molecule experiments became possible that monitor folding of the ubiquitin protein under constant force applied via an Atomic Force Microscope (AFM) and a long lived intermediate was observed. We investigated this intermediate state at atomic detail by Molecular Dynamics simulations. Our results suggest that in the intermediate state secondary structure and hydrophobic clusters are constantly reforming, thus indicating that the intermediate is 'molten-globule' like.
Folding experiments using AFM
In the AFM setup, ubiquitin is tethered between a surface and a nanoscale tip mounted at a flexible cantilever (Fig. 1a). After unfolding at a very high force (e.g. 100 pN), a weaker force of 20-40 pN is applied. At low forces, the resulting relaxed unfolded state (Fig. 1a, left) shows large distance fluctuations and eventually folds to the native state (Fig. 1a, right) as indicated by a sudden decrease in the end-to-end distance (graph in Fig. 1a). Thus, in contrast to previous ubiquitin folding studies, here a long-lived folding intermediate is observed, featuring nanometer scale length fluctuations prior to the final folding event.
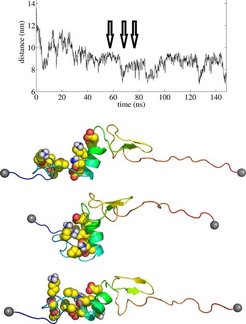
What is the molecular structure of this intermediate? And what are the forces and dynamics underlying the large fluctuations? We used Molecular Dynamics simulations to investigate the intermediate state at atomic detail. After partially unfolding ubiquitin under high force in the simulation, the force is quenched and the dynamics of the collapsed state on a several 100 ns scale are monitored in terms of the end-to-end distance, as in the AFM.
We find a very broad ensemble of non-native conformations, forming manifold hydrophobic and polar interactions. Secondary structure and hydrophobic clusters are of only temporary nature and are constantly reforming. No specific structure was found to be common within the ensemble. In this sense, the intermediate is 'molten-globule' like, here however stretched out by the external force.
The dynamics of the protein are associated to large scale end-to-end distance fluctuations similar in magnitude to the experiments. An example for the end-to-end distance found in our force-clamp MD simulations is shown in Figure 2. In a representative series of three conformations, fluctuations in the distance indicate clustering of hydrophobic residues, partially also formation of salt bridges, and their rupture shortly after. We interpret these fluctuations as folding attempts.
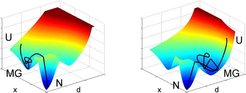
The similarity of the molten-globule like state found here with the non-native state of ubiquitin in solution studied so far suggests that the folding mechanisms underlying folding under force and temperature- or urea induced folding might share intermediates along the folding pathways (Fig. 3).
Future experiments allowing to monitor several folding variables in parallel might help to gain insight into the connection between the different reaction coordinates probed in experiments and simulations.


