The Primary Steps of the pH-Dependent Insulin Aggregation and the Role of Entropy
Collaboration: Andreas Bögehold, Avishay Pelah and Bernd Abel (University of Leipzig)
Insulin aggregation critically depends on pH. However, the underlying energetic and structural determinants are unknown. Applying combined mass spectrometry, atomic force spectroscopy, and molecular dynamics simulations, we have obtained structural information of putative aggregation rate-determining transition states.
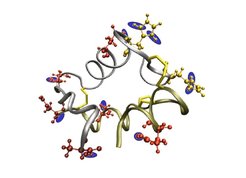
We measured the kinetics of the primary aggregation steps of the insulin monomer in vitro and related it to its conformational flexibility. To assess these primary steps the monomer concentration was monitored by mass spectrometry at various pH values and aggregation products were imaged by atomic force microscopy. Lowering the pH from 3 to 1.6 markedly accelerated the observed aggregation kinetics. The influence of pH on the monomer structure and dynamics in solution was studied by molecular dynamics simulations, with the protonation states of the titrable groups obtained from electrostatic calculations. Reduced flexibility was observed for low pH values, mainly in the C-terminus and in the helix of the B-chain; these corresponded to an estimated entropy loss of 150 J mol-1 K-1. The striking correlation between entropy loss and pH value is consistent with the observed kinetic traces in the experiment.
