Pause & go: How cells tune gene expression
In eukaryotes, the 12-subunit Pol II enzyme transcribes most protein coding genes. Pol II activity is carefully controlled to ensure the correct messenger RNA (mRNA) products are made. When Pol II is not properly controlled, diseases such as cancer can arise. Our group is primarily interested in uncovering the mechanisms cells use to regulate Pol II activity. Pol II is controlled as it first starts transcribing (initiation) and as it extends the RNA chain (elongation). Our group has elucidated many of the mechanisms regulating Pol II initiation in yeast (Schilbach et al., 2017; Hantsche and Cramer 2017). However, the mechanisms underlying mammalian elongation control have remained elusive, mainly due to the lack of structural information.
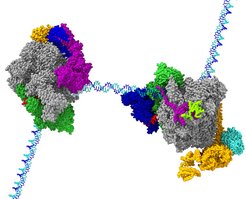
Shortly after Pol II has escaped from the gene promoter it often stops. This is referred to as “promoter-proximal pausing”. Pol II remains stably bound to the DNA, but does not progress further. Why does Pol II stop before it progresses to make the gene product? This process can be compared to a flutist in an orchestra. The flutist is always prepared to play but only plays when the conductor gives the proper cue. When Pol II stops in the promoter-proximal region, it behaves like the flutist. It waits until it receives the signal from the conductor or cell, to play or proceed. The cell uses elongation control, in addition to initiation control, to tune and orchestrate which gene products are produced at a particular time. Although the process of promoter-proximal pausing is essential for cell differentiation and organism development, and has been studied extensively by researchers world-wide, the mechanisms for Pol II pausing and for its activation have remained unknown.
In two studies, we now defined how specific factors stabilize paused Pol II and how other factors lead to its activation. We first reconstituted a paused transcription complex and determined its structure. Two factors, DSIF and NELF, are required for pausing Pol II. We prepared these factors recombinantly and established biochemical conditions where their function can be observed in vitro. Using cryo-EM, we were able to visualize Pol II on a DNA pause sequence from human immunodeficiency virus (HIV) bound to both DSIF and NELF. By following how Pol II, DSIF, and NELF associate with each other and with the DNA, we can now explain how Pol II pausing occurs and how it is stabilized. The underlying DNA sequence leads to a conformation of nucleic acids within the transcribing enzyme that impairs Pol II progression. NELF stabilizes Pol II in this non-productive state by restricting its mobility and by preventing positive factors from binding.
In the second study, we investigated how Pol II is released from the promoter-proximal region and how it is activated for rapid elongation. In an orchestra, the wave of the conductor’s wand signals the flutist to play. In cells, the signal to leave the promoter-proximal region comes from a machine that modifies Pol II, DSIF, and NELF by phosphorylation. Phosphorylation of the paused Pol II complex results in NELF loss and allows for additional factors to associate with Pol II. We investigated biochemically how the release of pausing is coupled to escape into rapid elongation and further identified the responsible factors. We found that phosphorylation by a specific enzyme complex called positive transcription elongation factor b (P-TEFb) allows for association of two factors, PAF and SPT6, with Pol II.
Function of PAF resolved
A key finding of this work resolves the function of an enigmatic protein complex called PAF. PAF acts as a switch between pausing and active elongation. It blocks NELF association, thereby converting Pol II from the paused state into a state that is competent for rapid elongation. DSIF, PAF, and SPT6 stabilize Pol II in a state that allows it to transcribe much faster than Pol II by itself. Using cryo-EM, we were able to see how DSIF, PAF, and SPT6 interact with Pol II to activate it. The factors coat the outer surface of Pol II. Comparison of the cryo-EM models from both studies shows that NELF and PAF occupy the same region on the Pol II surface. It also illustrates that DSIF adopts a different position in both complexes that may explain how DSIF can first support pausing and then activation.
Our studies combined biochemistry and cryo-EM. A key hurdle to our studies was to obtain pure protein factors. This obstacle was overcome by optimizing conditions to overexpress human protein complexes in insect cells. Using a combination of affinity purification and functional assays, we were then able to determine how well the factors associate with Pol II and how they affect Pol II activity. To visualize a sample by cryo-EM, it is placed on a small metal mesh made of copper or gold. The mesh is rapidly frozen to capture the sample in glass-like, vitreous ice. An electron beam is then passed through the sample to produce a 2D image with low contrast. To improve the signal and to get different views of the sample, hundreds of thousands of images are obtained. These images are then averaged and reconstructed in 3D space to determine a three-dimensional structure. The resulting three-dimensional maps can then be used to trace the amino acid backbone and side chains of the protein molecules or for docking crystal structures into matching densities. Modelling is aided by a technique called crosslinking mass spectrometry, which assists in identifying interacting regions of protein molecules. This technique was vital to our efforts and was performed in collaboration with the research group Mass Spectrometry headed by Henning Urlaub at the institute.
Together, our two new studies provide the basis for understanding how Pol II activity is controlled during transcription elongation in mammals. By visualizing the paused and activated states of mammalian Pol II, we have identified how different factors collaborate with one another and how they influence Pol II activity. This work provides a foundation for exploring how other factors contribute to these processes and how this type of regulation is used to tune cell type-specific gene expression.
References
Schilbach, S. et al.: Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017).
Hantsche, M. and Cramer P.: Conserved RNA polymerase II initiation complex structure. Current Opinion in Structural Biology 47, 17-22 (2017).












