Kinetic Simulation of Biosynthetic Reaction Networks Guided by Atomistic Free Energy Calculations
Collaborators: Grininger Lab, Goethe University Frankfurt, Germany

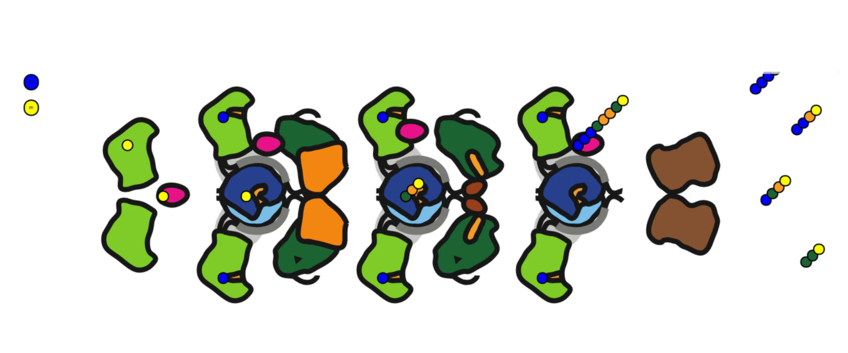
Fatty acid synthases (FAS) and polyketide synthases (PK) catalyse the synthesis of complex organic compounds through the interplay of a multitude of catalytic sites contained within multi-enzyme complexes. The fine control of product output observed in nature is achieved by enzymes and multi-enzyme complexes operating either iteratively or in series, with independent active sites responsible for priming, elongation and chemical modifications, subject to precise kinetic regulation to enable high product specificity from a collection of individual stochastic reaction processes. Achieving predictive understanding of the effects of specific perturbations necessitates techniques that can adequately represent the complex interplay and non-linear network effects that define the outputs of such networks.
We pursued a stochastic kinetic simulation approach to represent the reaction network of yeast typ I FAS, a synthetic megacomplex that incorporates six distinct catalytic activities in one macromolecule. Working with only sparse biochemical input data, we were able to search through the large reaction constant parameter space to obtain a sample of models that match the input data. In collaboration with the research group of Prof. Martin Grininger (Goethe University Frankfurt), we subsequently cross-validated the resulting ensemble model by incorporating calculated effects of active site mutations in accordance with free energy values calculated from atomistic molecular dynamics simulation, and were able to capture emergent non-linear features of the product output spectrum that had resisted explanation by conventional chemical intuition.
Having demonstrated posterior understanding of a well-studied system, we seek to apply the methodology to the targeted synthesis of custom polyketide chemicals.
