Electron-Spin Resonance Spectroscopy
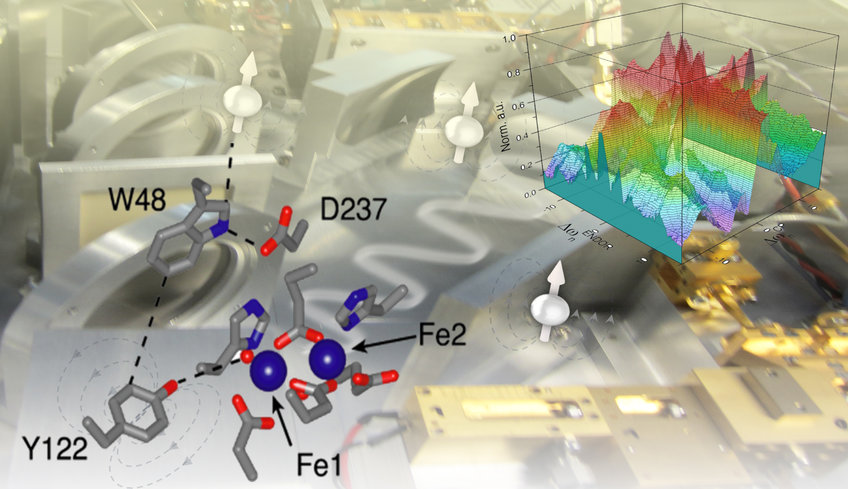
The research group is dedicated to modern electron-spin resonance (ESR/EPR) spectroscopy, from high-frequency EPR to methods at the interface with nuclear magnetic resonance (NMR) and their applications in biological science. We have been investigating how to excite and detect paramagnetic centers and their coupled nuclear spins with microwave (mw) and radio frequency (rf) pulses to achieve information on active sites of proteins or on the global structure of biomolecules. One main strategy is to transfer the much larger polarization of electron spins to nuclear spins. To this end, developments of pulse schemes at high magnetic fields (≥ 3 Tesla) and corresponding resonant mw frequencies are in focus. The most representative applications are the investigation of enzymatic reactions involving paramagnetic intermediates, particularly the proton-coupled electron transfer (PCET) in E. coli ribonucleotide reductase (RNR), and the exploration of long-range structural information in nucleic acids and transmembrane peptides by pulsed dipolar spectroscopy.
Press releases & research news
