Switching on genes for energy production
Humans, animals, and plants all depend on oxygen to live. They utilize oxygen to produce energy in a process called respiration, which takes place in specialized cellular organelles, the mitochondria. These “powerhouses” can be found in nearly every cell in our body. They are thought to have arisen around 1.5 billion years ago, when an evolutionary ancestor of our modern-day cells engulfed a bacterium. This marked the beginning of a productive symbiosis that has lasted to the present day and is thought to be the originating event of the eukaryotic lineage. As an evolutionary remnant, mitochondria contain a separate DNA genome as well as the molecular machinery necessary to replicate it and to express its genetic information.
Although the human mitochondrial genome is small compared to the nuclear genome, the genes encoded by it are essential. Without them, respiration cannot take place. Thus, activation of these genes must be controlled by the cell in response to varying energy demands. The first step in activating mitochondrial genes consists of gene transcription, when the genetic information encoded in the DNA is copied into RNA. Transcription is carried out by the mitochondrial RNA polymerase, which is evolutionarily neither related to polymerases found in the cell nucleus nor polymerases in bacteria.
Several years ago, our research group reported the first structures of the mitochondrial RNA polymerase, both in free form (Ringel et al., Nature 2011) and with bound DNA template and RNA transcript (Schwinghammer et al., NSMB 2013). These studies revealed how the enzyme resembles polymerases used by bacteriophages. However, they did not show how mitochondrial genes are transcribed because they did not include the additional protein factors that the polymerase requires to exert its function. In particular, two protein factors – called TFAM and TFB2M – are required for initiation, and one factor – TFEM – is required for RNA chain elongation and ensures that transcription is not prematurely terminated. It thus remained unknown how these three factors enable the mitochondrial RNA polymerase to fulfill its function.
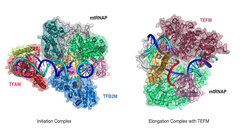
Now, in a collaboration with the laboratory of Dmitry Temiakov at Rowan University, Stratford (United States), we have arrived at an understanding of factor-dependent mitochondrial transcription. In a first study, we aimed to understand how the mitochondrial RNA polymerase can initiate transcription. We could visualize the initiation complex formed by polymerase on DNA with the two initiation factors TFAM and TFB2M by X-ray crystallography. The structure explains how TFAM can recruit the polymerase to DNA and how it positions it at the beginning of the gene. The second step during initiation consists of DNA opening by separating its two strands, so that the sequence information can be read. This is where TFB2M comes into play. TFB2M changes the structure of the polymerase to position one of its elements such that DNA opening occurs. TFB2M also binds one of the DNA strands to prevent the DNA duplex from closing again.
Now, RNA synthesis begins and the initiation factors dissociate from the polymerase, which proceeds to the elongation phase. To complete transcription, the polymerase must remain stably bound to DNA until the end of the gene. This is where the elongation factor TEFM is required. In its absence, the polymerase terminates transcription prematurely, leading to a short RNA molecule that serves as a primer for DNA replication. The interaction with TEFM, however, enables the polymerase to transcribe stably, and to complete the RNA message. Hence, TEFM is a switch that regulates whether short RNA is produced and mitochondrial DNA is replicated or whether long RNA transcripts are made and mitochondrial genes are transcribed. How TEFM achieves this remained unknown.

In a second study, we collaborated with the Research Group of Bioanalytical Mass Spectrometry headed by Henning Urlaub at the MPI-BPC to decipher how TEFM assists the polymerase to stimulate elongation. We first determined the three-dimensional structure of TEFM at atomic resolution. Then, we visualized the elongation complex consisting of polymerase, TEFM, DNA, and RNA. This structure reveals that TEFM clamps the DNA and RNA to stabilize the elongation complex and to prevent the formation of secondary structure in the nascent RNA transcript that can present obstacles during transcription.
Taken together, a combination of structural and functional studies provided the fundamental mechanisms underlying gene transcription in human mitochondria. These new studies led to a molecular movie of this important cellular process that depicts the roles of the three mitochondrial transcription factors during polymerase action. Mitochondrial transcription is found to be an evolutionarily unique system that differs substantially from transcription in the cell nucleus or in bacteria. Our new data nevertheless suggest that these distinct transcription systems share conceptual similarities and follow general mechanistic principles.
Hauke S. Hillen and Patrick Cramer
References
Schwinghammer, K., Cheung, A. C. M., Morozov, Y. I., Agaronyan, K., Temiakov, D., & Cramer, P. Structure of human mitochondrial RNA polymerase elongation complex. Nature Structural & Molecular Biology 20, 1298–1303 (2013).
Ringel, R., Sologub, M., Morozov, Y. I., Litonin, D., Cramer, P., & Temiakov, D. Structure of human mitochondrial RNA polymerase. Nature 478, 269–273 (2011).












