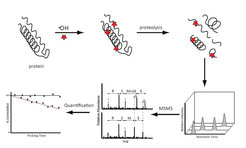
Structural Probing of protein and protein(-ligand) complexes
The identification of interaction surfaces within macromolecular complexes is a major goal in the bioanalytical sciences. We attempt a combined approach of oxidative footprinting and chemical labeling that searches to shed light on protein-protein and protein-ligand interaction sites. Oxidative footprinting allows to map the surface of a protein or protein complex, as it is covalently modified, leading to a variety of oxidation products. The high reactivity of the hydroxyl radical allows for a non-selective, widespread exploration of a protein’s surface. We have developed a data-analysis strategy that involves the identification and label-free quantification of modified peptides. In this way, we can map the surfaces of the proteins and protein complexes of interest and obtain results in good agreement with solvent-accessible areas calculated from crystal structures. In addition, we can identify residues involved in interaction at protein–RNA and protein–protein interfaces.