New tools for research on mammalian oocytes
To ensure that we build a solid foundation for future research in our laboratory, we successfully developed new technology to study meiosis in mammalian oocytes. For instance, we developed an experimental strategy to carry out high-content screens in mouse oocytes; and we have been able to develop techniques that allow us for the first time to study meiosis in live human oocytes.
Live imaging screen reveals causes of meiotic defects in mammalian oocytes
To understand why human oocytes are so frequently abnormal we need to identify the genes that safeguard accurate progression through meiosis. To achieve this aim, we developed an experimental strategy that allowed us to carry out the first RNAi screen in mammalian oocytes. We analyzed the function of 774 genes by high-resolution imaging of chromosomes and microtubules during meiosis in live mouse oocytes and scored each oocyte quantitatively for 50 phenotypes, generating a comprehensive resource of meiotic gene function (Fig. 1). This resource allowed us to identify and characterize many new genes essential for meiosis in mammalian oocytes. The screen generated an unprecedented annotated dataset of meiotic progression in 2,241 mammalian oocytes. This dataset allowed us to analyze systematically how meiotic defects arise (Pfender et al., 2015).

Figure 1. Scheme illustrating principle of RNAi screen in mouse oocytes. siRNAs targeting several genes simultaneously were microinjected into follicles, which were then cultured in vitro for 10 days. Chromosomes and microtubules were imaged and phenotypes were assessed in more than 2,200 live oocytes.
Rapid degradation of endogenous proteins by Trim-Away
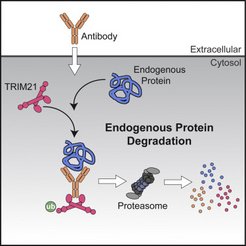
Methods for the targeted disruption of protein function have revolutionized science and greatly expedited the systematic characterization of genes. Two main approaches are currently used to disrupt protein function: DNA knockout and RNA interference, which act at the genome and mRNA level, respectively. A method that directly alters endogenous protein levels is currently not available. We developed Trim-Away, a technique to degrade endogenous proteins acutely in mammalian cells, including oocytes, without prior modification of the genome or mRNA. Trim-Away harnesses the cellular protein degradation machinery to remove unmodified native proteins within minutes of application. This rapidity minimizes the risk that phenotypes are compensated and that secondary, non-specific defects accumulate over time. Because Trim-Away utilizes antibodies, it can be applied to a wide range of target proteins using off-the-shelf reagents. Trim-Away allows the study of protein function in diverse cell types, including non-dividing primary cells where genome- and RNA-targeting methods are limited (Clift et al., Cell 2017)
References
Pfender, S., Kuznetsov, V., Pasternak, M., Tischer, T., Santhanam, B., and Schuh, M. (2015). Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature 524:239-42.
Clift, D., McEwan, W., Labzin, L.L., Konieczny, V., Mogessie, M., James, L.C., and Schuh, M. (2017). A method for the acute and rapid degradation of endogenous proteins. Cell, doi: 10.1016.



