Start me up – initiation of protein production in human cells filmed
Max Planck researchers uncover in molecular detail how the translation machinery selects the start site of a protein-coding region on mRNA for protein production in human cells.
All life depends on finely tuned protein production. In human cells, an intricate molecular machinery called 48S translation initiation complex plays a key role in regulating which genes are translated into a protein. A team led by Niels Fischer and Marina Rodnina at the Max Planck Institute (MPI) for Multidisciplinary Sciences in Göttingen (Germany) has now reconstituted the complete 48S machinery in the test tube and succeeded in “filming” how the translation machinery selects the start site of a protein-coding region on mRNA for protein production in molecular detail. This reveals how structural changes ultimately define the set of proteins in a human cell.
All cells in our body contain exactly the same genetic information, which is encoded in the genes in our DNA. But different cell types activate only the genes they need to perform their respective tasks. Therefore, tight regulation of gene expression is essential for living cells in order to vary their set of proteins. For a long time, gene expression was thought to be controlled mainly at the level of transcribing DNA into messenger RNA (mRNA). However, researchers over the past two decades have shown that the subsequent step, the regulation of translation of mRNA into proteins, can be equally important. Translation is generally regulated during its initiation phase, but the complexity of initiation in human cells has hindered a detailed molecular understanding of this fundamental process so far.
A complex Lego brick system for structural studies

To observe remodeling of the complex 48S machinery (48S) on the molecular level, the MPI researchers purified the numerous and fragile components of the 48S machinery, including a dozen of initiation factors (eIFs) and the small ribosomal subunit, from human cell culture. “Our approach can be compared to a sophisticated Lego brick system,” co-author Sung-Hui Yi from Rodnina’s Department of Physical Biochemistry explains. “After purifying the individual components, we were able to assemble the complete human 48S machinery under well-controlled conditions in the test tube. This was essential for the next step: visualizing the manifold changes in the 48S structure that are key to its function by cryo-electron microscopy (cryo-EM).”
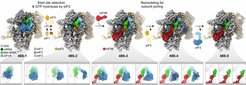
To this aim, the researchers had to resolve the resulting mixture of 48S complexes in different states of the initiation process by extensive computational analysis of a large cryo-EM data set. “A daunting task,” according to first author Valentyn Petrychenko from Fischer’s project group Molecular Machines in Motion, “because we had to acquire more than six million cryo-EM images and sort them computationally to visualize the full sequence of structural changes by the 48S machinery.” The obtained snapshot series of key 48S intermediates provides a ‘molecular movie’ that reveals in detail how the translation machinery selects the start site of a protein-coding region on mRNA for protein production.
Finding the right start
After binding to the mRNA, the 48S moves along the mRNA to scan for start sites, very much like a bar code scanner. Groundbreaking work more than four decades ago showed that in eukaryotes the efficiency of start site selection, and, thus, the efficiency of protein production, is regulated by a specific mRNA sequence called Kozak sequence, which flanks the universally conserved AUG start codon.
“Excitingly, our structures now unravel the molecular mechanisms by which the Kozak sequence regulates start site selection and how the subsequent hydrolysis of a GTP molecule commits the ribosome to the start site,” explains Max Planck Director Rodnina. The researchers found that the Kozak sequence first ‘slows down’ the scanning 48S machinery and then ‘stops’ it by stabilizing local and global structural changes of the 48S upon start codon recognition. As a consequence, the 48S-bound protein complex eIF2 hydrolyzes GTP, an irreversible chemical step, that triggers a dramatic remodeling of the 48S complex in both structure and composition.
Final check by the large eIF3 protein complex
By visualizing this multi-step remodeling process, the scientists could follow how it completes start site selection and how it prepares the 48S for the final step of initiation, the joining of the large ribosomal subunit to form a complete ribosome ready for protein production. “We discovered that the large multi-protein eIF3 complex can sense the state of 48S remodeling to precisely control the timing of subunit joining, making sure it only happens, when the entire 48S machinery is ready,” says project group leader Fischer.
“Errors in translation initiation can lead to diseases such as cancer and mental disorders,” Fischer adds. “Therefore, the present study not only reveals how the 48S machinery starts protein production, but also provides a basis for future studies on how dysregulation of the initiation process drives disease.” (nf)










