ENDOR for information from the atomic to nano scale
Electron-nuclear double resonance (ENDOR), particularly at high frequencies, permits to determine the distance as well as the orientation of nuclear spins in the ligand sphere (r < 1.5 nm) of a paramagnetic centre. For instance, hydrogen bond interactions in the active states of an enzyme can be detected that are not easily accessible by other techniques such as X-ray, cryo-EM or NMR. The combination of ENDOR with quantum chemical calculations becomes a unique tool to elucidate the structure of complex intermediates during enzymatic catalysis. We have recently demonstrated the spectroscopic detection of hydrogen bond networks in proteins via 2H ENDOR. The 17O detection of water molecules participating in proton-coupled electron transfer (PCET).
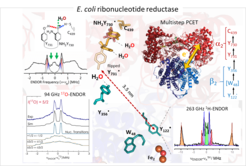
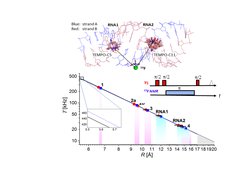
19F ENDOR for distance measurements: Fluorine labelling has been established in NMR as powerful and selective tool to extract structural information in biological systems. 19F can also be detected in ENDOR by taking advantage of the large electron spin polarization. We have recently shown that 19F ENDOR spectroscopy can be used to complement EPR distance measurements in the angstrom to nanometer range by introducing a nitroxide and a 19F in the molecule of study. This can be performed in nucleic acids as well as in enzyme and proteins. New labelling procedures to site-selectively introduce unnatural amino acids in proteins open the route for this type of studies. Currently, we are employing this method to address the open question of the PCET mechanism in RNR across the subunit interface. Measurement of the distance across the subunit interface in active E.coli RNR will provide important new structural information.
Development of ENDOR pulse sequences (DFG SPP-1601): In the past year, we have started a detailed investigation of ENDOR pulse sequences to optimize their performance for different interaction ranges and different nuclear spins. We are examining the effect of nuclear spin relaxation and proposing new sequences based on electron spin lock and electron-nuclear cross polarization, to circumvent signal saturation effects. We have been implementing analytical and numerical tools to predict ENDOR signals based on a static spin Hamiltonian as well as density matrix formalism, product and single transition operators (Bejenke et al, Mol. Physics 2020). The goal is to predict the efficiency of more complex pulse sequences in powder patterns under real sample conditions, i.e. low temperatures and high electron spin polarization.
Statistical methods for ENDOR (DFG-CRC 1456): Mathematical methods are gaining increasing importance in various fields of spectroscopy to extract weak signal from noise and baseline distortions. This applies also for ENDOR spectra, where resonances are often very broad and close to the baseline. In order to address this issue, we have started a collaboration with the group of S. Huckemann (Dept. Mathematical Stochastics, Göttingen) and Yvo Pokern (UCL London) focusing on statistical analysis of ENDOR data. This approach allows for the separation of signal drifts during long acquisition times and for the determination of the most-likely ENDOR signal with uncertainties, paving the way for more quantitative data analysis (Pokern et al., PNAS DOI 10.1073/pnas.2023615118).

