Dealing with myelin differences in mice and humans
If the myelin layers of neuronal axons become damaged, the electrical impulses travelling along the neurons slow down, affecting the ability to walk, speak, see, or think. This can cause several illnesses, including multiple sclerosis and a group of rare genetic diseases known as leukodystrophies.
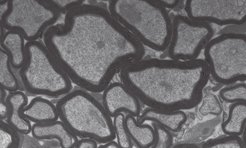
Human myelin disorders are commonly studied in mouse models. However, due to the many obvious physical differences between men and mice, it is important to determine whether the results of experiments performed in rodents are applicable to humans. To do this, it is necessary to understand how myelin differs between the two species at the molecular level.
We approached this problem by studying the proteins found in myelin isolated from the brains of body donors. With quantitative mass spectrometry, we produced a list of proteins in human myelin that could then be compared to existing data from mouse myelin. This analysis showed that myelin is very similar in both species, yet some proteins only appear in humans, some only in mice. We also compared which genes are turned on in the oligodendrocytes producing the myelin.
It became evident that there are unexpected differences between the myelin of humans and mice that will need to be considered when applying results from mice research to humans. We therefore created a web interface to enable the access to our data on the proteins in myelin and the genes expressed in oligodendrocytes in the two species. To make our data permanently available in an interactive way, we collaborated with the Gesellschaft für wissenschaftliche Datenverarbeitung Göttingen (GWDG) which hosts the data sets and the necessary applications on its servers. Within a short period of time, the GWDG has implemented the technical and legal requirements for our project. Thanks to this open science solution, scientists can now access relevant data with just one search request without getting into the depths of an extensive data repository.
To the Proteome and Transcriptome Data Access




