Like a wedge in a hinge
Researchers lay groundwork to tailor drugs for new targets in cancer therapy
Thanks to medical advances, many types of cancer are treatable nowadays. However, a panacea for cancer is still a long way off. With some cancer types the available therapies reach their limits because either the tumor does not respond to the treatment from the onset or it becomes resistant after some time. Scientists are therefore developing strategies to tackle cancer cells at spots that have not been the target of drugs so far.
Such a clinically largely untested starting point is the protein complex SF3B. It is instrumental in the first steps of the production of proteins, the universal tools of living cells. To produce proteins, the cell first needs to bring the protein blueprints into a readable form. To this end, the blueprints are cut and recombined in a sophisticated process by a complex molecular machine, the spliceosome. SF3B, as part of the spliceosome, controls at which point the building instructions are cut. If errors occur in this step, the cell produces altered proteins which might severely disrupt cellular processes.
A printable version of the image may be downloaded here.
The idea of the researchers: They want to manipulate the function of SF3B and thus mess up the production of certain proteins in order to kill cancer cells. Scientists were already able to develop agents that bind to SF3B. These do not block SF3B completely but modulate its function, with the result that some protein blueprints are cut differently. These alterations affect cancer cells more than healthy cells.
“However, so far we know very little about how exactly these substances interact with SF3B,” says Vlad Pena, who heads the Research Group of Macromolecular Crystallography at the MPI for Biophysical Chemistry. “But this information is essential to improve the agents so that they may serve as anti-cancer drugs.”
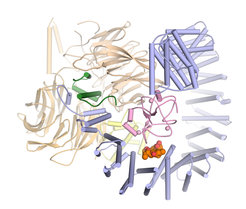
In collaboration with the pharmaceutical company H3 Biomedicine Pena’s team has now taken a decisive step: “For the first time, we were able to determine the three-dimensional structure of SF3B in interaction with an active substance in atomic resolution,” the structural biologist relates.
Valuable insights for drug optimization
The scientists’ results reveal in detail how the active substance pladienolide B attaches to SF3B and interferes with its function. “Pladienolide B acts like a wedge in a hinge and prevents SF3B from pivoting. This movement is necessary for SF3B to function reliably,” explains Constantin Cretu, a researcher in Pena’s team and first author of the study now published in the journal Molecular Cell.
The new insights explain previous results on similar active substances, because pladienolide B is representative of a whole class of chemical agents that vary greatly in their form but share one important feature: They all have the same chemical group in their center. “Until now, it was unclear why this chemical group is so important,” Cretu says. “Our structure of SF3B and pladienolide B now shows that precisely this group substantially contributes to the binding of the drug and related substances to SF3B.”
Moreover, the researchers’ data maps all further contacts between pladienolide B and SF3B. Based on these data one can predict where the drug can be modified and where not, Pena points out: “We hope that our insights will serve as a guide to developing novel anti-cancer agents in the future.” (fk)












