Ribosomes shifting the reading frame
Ribosomes are ancient molecular machines that produce proteins in the cells in a process called translation. The genetic information encoded in the messenger RNA (mRNA) is read by the ribosome in a triplet format termed codon. The sequence of codons in the mRNA defines the sequence of amino acids in the proteins. Therefore, the exact triplet-wise step of decoding has to be maintained by sophisticated mechanisms. While maintenance of the correct reading frame is generally very accurate, in specific cases alterations of decoding rules become advantageous. In some cases, the reading frame may shift in -1, +1, or even in -2 directions in a programmed fashion. Normally, erroneous frameshifting leads to production of a wrong peptide or to premature termination. Programmed frameshifting, however, provides the ribosome with an alternative reading frame for translation and leads to synthesis of functional proteins. The efficiency of programmed frameshifting can be as high as 50%, which implies that half of the ribosomes move to a new reading frame upon reaching a specific signal embedded in the mRNA. At the end, the final translation products of such an mRNA are two proteins which have identical N-terminal sequences but are different in the C-terminal parts.
This mechanism is especially useful for viruses which have a limited genome size, because it expands their genetic repertoire. Many viruses which possess RNA as the genetic material, including HIV and SARS, commonly use programmed frameshifting to express the genes critical for their proliferation and viability. This makes frameshifting a very attractive target to design anti-viral drugs, which has been exploited for a decade. However, the mechanism of programmed frameshifting remained obscure. This prompted Marina Rodnina’s research team of the Department of Physical Biochemistry at the Max Planck Institute for Biophysical Chemistry to inquire how exactly the ribosomes slip on the message to change the reading frame.
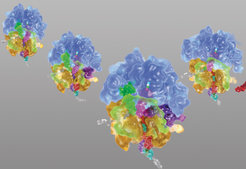
Efficient frameshifting depends on orchestrated action of multiple signals embedded in the mRNA. In case of -1 frameshifting, one of these elements is a slippery sequence, usually in the form of a heptanucleotide stretch of nucleotides such as UUU AAA G. This is the sequence on which the ribosome can change the reading frame. The second element is a stimulatory structure, which can be found in the form of a hairpin or a more complex pseudoknot structure. Upon encountering these structures, ribosomes pause, because their movement over the mRNA is hindered by the bulky structures (Figure 1).
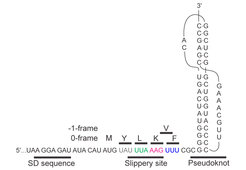
Taken from: Neva Caliskan, Vladimir I. Katunin, Riccardo Belardinelli, Frank Peske, Marina V. Rodnina: Programmed –1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619-1631 (2014).
Monitoring -1 frameshifting in real time
To understand when exactly the slippage occurs, a so called codon walk was performed over the frameshifting sequences, i.e. ribosomes were monitored moving one codon at a time. This was only made possible by using a purified biochemical system from bacteria composed of translation components such as ribosomes, elongation factors powered with GTP, frameshifting mRNAs, and various aminoacylated tRNAs. At each step, the relative amount of ribosomes moving in the 0- or -1 frame was measured. With the native slippery sequence and the stimulatory pseudoknot, frameshifting took place in 70% of the cases, which is in agreement with the cellular levels of frameshifting determined in bacterial reporter assays. The ribosome changed the frame after reading both slippery codons, but before decoding of the codon following the slippery site. Thus, the slippage occurred exactly at the moment when the two tRNAs together with the mRNA had moved through the ribosome in a process known as translocation.
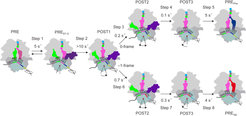
Impeded translocation during -1 frameshifting
Translocation of tRNAs and mRNA in bacteria is facilitated by a protein called elongation factor G (EF-G). Translocation is a very dynamic process which involves the rotation of ribosomal subunits relative to each other as well as complicated motions within the small (30S) subunit. Because usually the ribosomes translocate exactly by one codon, the question is why and when they start to slip. To answer these questions, we placed fluorescent reporters on the ribosome, tRNAs and EF-G, and tracked the kinetics of the individual movements. Specifically, we looked at binding and dissociation of the EF-G, movement of the tRNA through and out of the ribosome and the movement of the 30S subunit. The initial phases of translocation were not affected: EF-G could bind to the ribosome, and promote rapid forward movement of the tRNAs together with the 30S subunit. However, at the end of translocation, the 30S subunit has to move backwards, which would result in a netto movement of the ribosome on the mRNA by one codon. Exactly this backward movement was impaired by the pseudoknot: The strong secondary structure on the mRNA provided a steric challenge to the ribosome and impeded the forward movement. The steric clash was resolved if the ribosome shifted over the slippery sequence into the -1 frame. Apparently, this placed the base of the pseudoknot into the helicase active site of the ribosome, thereby allowing for more efficient unwinding of the secondary structure and moving along the mRNA. In kinetic terms, frameshifting can be envisaged as a choice between two alternatives, the very slow reading in the 0 frame versus faster movement in the altered, -1 frame (Figure 2). In the cell, this decision is encoded in the structure of the slippery site and the mRNA secondary structure element, thereby providing the desired ratio between the two alternative protein products. Thus, the mRNA not only contains the information for the sequence of amino acids, but also defines how much of an alternative product will be produced. One additional unexpected outcome of our work is the finding that the slippery sequence alone can stimulate very efficient and rapid movement into the -1 reading frame. This implies that recoding events on the slippery sequences in the genomes might happen more often than so far reported. If true, this would provide an additional, yet unappreciated source of protein diversity in the cell which might be used for adaptation to a changing environment. Understanding the mechanism of -1 frameshifting therefore is of crucial importance for understanding viral proliferation strategies in the host cells, as well as regulation of gene expression at the translational level.


