Elevating protein structure search to a new level
The rapid increase in publicly available protein structures, due to advancements in protein structure prediction methods like AlphaFold2 and ESMFold, has led to unprecedented growth of protein structure databases. Since structures are much more conserved than protein sequences, detecting distant evolutionary relationships works much better when comparing structures than when comparing their sequences. The vast structure databases thereby in principle massively improve researchers’ ability to find distantly related proteins. This allows them to learn the functions of proteins based on already studied relatives.
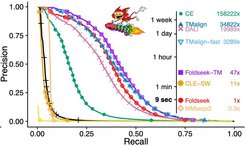
Making efficient use of this treasure trove of structural data requires frequent searches of these databases for structures similar to proteins of interest. However, existing search methods would take weeks or months for a single search with a protein of interest to find similar structures among the 200 million structures in the current databases.
To address this computational challenge, researchers from Seoul National University (South Korea) and the Max Planck Institute for Multidisciplinary Sciences in Göttingen (Germany) have developed Foldseek, a groundbreaking protein structure search tool. This innovative tool reduces the search time from weeks to a mere few seconds while maintaining search sensitivities only slightly below the most sensitive current tools. To reach such high speed, Foldseek relies on a simple trick: Proteins are chains of chemical units called amino acids that fold into a stable 3D structure. Foldseek describes the structure as a sequence of letters, each of which encodes the 3D-interaction of one of its amino acids with its 3D-nearest neighbor amino acid. Foldseek then uses very fast sequence search tools to compare sequences instead of directly comparing their 3D structures.
With its superior speed and sensitivity, Foldseek leverages the enormous potential utility of the huge new protein structure databases for diverse life science fields such as molecular biology, molecular medicine, and microbiology. With a range of features, a webserver (search.foldseek.com), workflows and additional functionality, Foldseek enhances the protein structure search and alignment process, allowing users to customize their workflows and adapt Foldseek to their research needs. In conclusion, Foldseek increases the speed for searching similar protein structures around a factor of hundred thousand, making it an invaluable tool for future structure-based analyses. For more information, refer to the recent publication in Nature Biotechnology.
