How RNAs are made-to-measure to power our cells
Research news
Mitochondria are organelles within our cells that generate the majority of chemical energy required to sustain life. They contain a dedicated genome with a special set of genes that need to be expressed to fulfill their function. An important step in mitochondrial gene expression is RNA processing, during which the premature RNA is cut into functional entities by specialized RNA-cutting enzymes. Defects in this process lead to severe human diseases, but the underlying molecular mechanisms are not well understood. Scientists at the Max Planck Institute (MPI) for Biophysical Chemistry and the University Medical Center Göttingen (UMG) have now determined the molecular mechanism of the first step of RNA processing in human mitochondria. This step is carried out by an enzyme complex called mitochondrial ribonuclease P (mtRNase P). The scientists visualized the structure of this molecular RNA cutting machine using cryo-electron microscopy (cryo-EM). This reveals for the first time how mitochondrial RNAs are cut, and explains why mutations in the involved proteins lead to disease in humans. The structure also provides clues as to why mitochondria have evolved a unique RNA-cutting machine that differs from those found in the cell nucleus or in other organisms.

Human cells contain two genomes. In addition to the well-known nuclear genome, a second genome exists in mitochondria. These organelles and their genome likely originate from certain bacteria, which were taken up by evolutionary ancestors of our cells millions of years ago. Today, they are the powerhouses of the cell, because they consume oxygen to produce adenosine triphosphate (ATP). ATP is the universal biological energy currency that fuels all biochemical reactions. The genes encoded in the mitochondrial genome are essential for this process, and mitochondrial gene expression is therefore crucial for cellular function.
In the first step of gene expression, the genetic information stored in the mitochondrial DNA is copied to RNA in a process called transcription. The mitochondrial transcription apparatus is different from its counterpart in the nucleus, and researchers Hauke Hillen and Patrick Cramer at the MPI-BPC have previously studied it in detail (Bonekamp et al., 2020; Hillen et al., 2017a, 2017b). One particularly intriguing difference is that, in mammals, mitochondrial genes are not transcribed individually gene by gene. Instead, almost the entire genome is copied into one long RNA molecule, which contains ribosomal RNA sequences and messenger RNA sequences (mRNAs) separated by transfer RNA (tRNA) sequences. To become functional, this so-called polycistronic RNA therefore needs to be cut precisely to release the individual RNA types. This critical step is known as RNA processing.
Human mitochondria have a specialized molecular cutting-machine, called mitochondrial ribonuclease P (mtRNase P). It recognizes the tRNA regions in the RNA and then cuts precisely at their beginning. Similar enzymes can be found in all domains of life. However, mtRNase P is different because it is a complex composed of three proteins of which only one resembles other known RNase P enzymes. The other two subunits are unique to mammalian mtRNase P. Moreover, mtRNase P not only cleaves the RNA but also chemically modifies it by adding a methyl group at a certain position, making it a multi-functional molecular machine. Malfunction of mtRNase P leads to severe mitochondrial disease in humans. Yet, the underlying precise mechanisms on how mtRNase P works as well as why mammalian mitochondria have evolved such a highly specialized and unique machinery were unknown.
First high-resolution structure of the human mitochondrial RNase P
Researchers at the MPI for Biophysical Chemistry and at the UMG have now deciphered how mtRNase P cuts RNA in human mitochondria. To achieve this, the team led by group leader Hauke Hillen first reconstituted the complex in a test tube and then determined its three-dimensional structure using cryo-EM. For this technique, the sample is rapidly flash-frozen in a thin layer of glass-like ice before it is imaged in an electron microscope at liquid nitrogen temperature. Because the molecules are randomly oriented in the ice, the resulting images represent two-dimensional projections of the sample from different angles. These images can then be averaged and used to reconstruct the three-dimensional structure of the target molecule at sub-nanometer resolution.
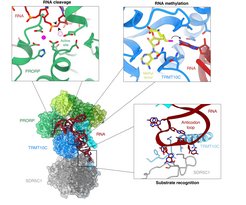
This approach allowed the researchers to obtain the first high-resolution three-dimensional structure of human mtRNase P. It shows how the three protein subunits bind to each other and to the mitochondrial RNA. The surprising finding: Mitochondrial RNase P interacts with a different part of the RNA than all other known RNase P enzymes. While these enzymes typically recognize a specific structure of the tRNA (called the ’elbow’), mtRNase P instead binds an element on the opposite side of the tRNA (the so-called anti-codon loop). “This was really unexpected at first. But it makes a lot of sense!”, Hillen says. “The mitochondrial genome in mammals has diverged strongly from its bacterial ancestor over the course of evolution, and mitochondrial tRNAs often differ from canonical tRNAs in their elbow structure”. The anti-codon loop, in contrast, is present in all mitochondrial tRNAs because it is required for their primary function: to interact with mRNAs at the mitochondrial ribosome. The interaction with the anti-codon part of the mitochondrial tRNA is mediated by the mtRNase P subunits unique to mitochondria. “This could explain why human mitochondrial RNase P is different from other RNase P enzymes and has additional subunits: It has to recognize different structures in the target RNA,” adds Arjun Bhatta, PhD student in Hillen’s group and first author of the study.
The structure also explains how mtRNase P combines two enzymatic functions – RNA processing and methylation – into one molecular machinery, and provides the molecular mechanisms for these functions. The two unique mtRNase P subunits are so-called moonlighting enzymes. This means they have multiple functions and carry out other tasks in the cell in addition to RNA processing. One of them (SDR5C1) is a dehydrogenase, a type of enzyme typically involved in metabolism, and the other one (TRMT10C) is a methyltransferase, an enzyme that adds a small chemical modification to RNA. The structure shows that the methyltransferase subunit acts as both an enzyme and a structural scaffold. Together with the dehydrogenase subunit, it forms a ’platform’ to which the RNA can bind. This positions it in a way that methylation can occur precisely at the correct position. It allows binding of the third subunit, which performs the cutting of the RNA. “This kind of re-purposing of apparently functionally unrelated protein folds seems to be a common theme in mitochondrial gene expression, as we had previously discovered similar adaptions in the mitochondrial transcription machinery,” says Hillen.
New insights into disease mechanisms
The detailed analysis of the human mitochondrial RNA cutting machine also provides insights into the molecular basis of some mitochondrial disorders. Genetic studies have shown that mutations in the subunits of mtRNase P lead to mitochondrial diseases in patients. The structure now allowed the researchers to precisely locate these known mutations within the mtRNase P. This shows that some mutations affect parts of the complex that are directly involved in RNA binding, and therefore likely lead to a decreased function of the RNA cutting machine. Thus, the structure provides a molecular explanation for how these mutations lead to diseases in humans.
In the future, Hillen and his team want to build on these results and unravel the mechanisms underlying the next steps of RNA processing in human mitochondria. (hh)
References
Bonekamp, N.A., Peter, B., Hillen, H.S., Felser, A., Bergbrede, T., Choidas, A., Horn, M., Unger, A., Lucrezia, R.D., Atanassov, I., et al. (2020). Small-molecule inhibitors of human mitochondrial DNA transcription. Nature 588, 712–716.
Hillen, H.S., Parshin, A.V., Agaronyan, K., Morozov, Y.I., Graber, J.J., Chernev, A., Schwinghammer, K., Urlaub, H., Anikin, M., Cramer, P., et al. (2017a). Mechanism of Transcription Anti-termination in Human Mitochondria. Cell 171, 1082-1093.e13.
Hillen, H.S., Morozov, Y.I., Sarfallah, A., Temiakov, D., and Cramer, P. (2017b). Structural Basis of Mitochondrial Transcription Initiation. Cell 171, 1072-1081.e10.












