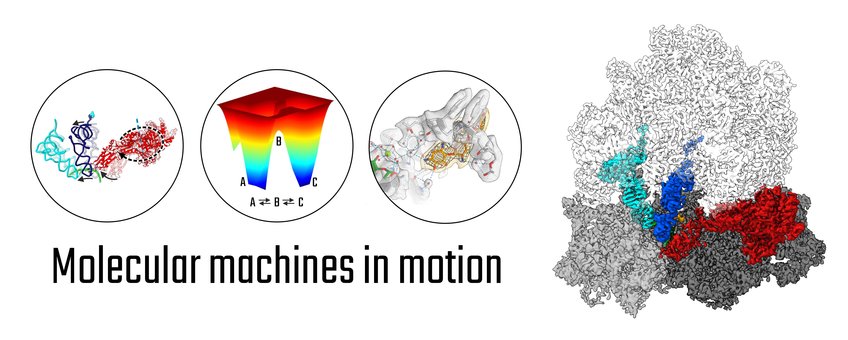
All organisms rely on molecular machines that undergo constant motion to perform their functions within their cells. The major question driving our research is to understand how these thermally-driven nanomachines work in health and disease. To this end, we advance and apply single particle cryo-electron microscopy (cryo-EM) to visualize the architecture and structural transitions of these machines at highest possible resolution. We are particularly interested in the structural dynamics of the ribosome, the protein-synthesizing factory of the cell, and how its dynamics can be modulated by antibiotics.
Conformational landscapes - Due to their microscopic environment, molecular machines work based on drastically different principles than their macroscopic counterparts. In particular, they use minor external energy to bias random thermal fluctuations into directed, large-scale conformational changes. Thus, to understand how these nanomachines work, it is essential to map their structural transitions – their conformational landscape – at highest detail at physiological temperatures. We use and advance time- and temperature-resolved cryo-EM approaches towards “filming” the full working cycle of macromolecular machines.
Molecular mechanisms and antibiotic action – How can molecular machines work at high speed and fidelity; how do they transmit signals? To uncover the working principles of molecular machines, we mainly focus on the ribosome, the prototype machine of gene expression. The ribosome is a large RNA-protein complex, which translates the genetic information encoded in mRNA into the amino acid sequence of a protein. We visualize the conformational landscape of protein synthesis by the bacterial ribosome to reveal the principles of molecular recognition and movement that enable the high speed and fidelity of the ribosomal machinery. We resolve ribosome-antibiotic complexes in functional states of translation to understand antibiotic action and resistance mechanisms and to provide a structural basis for improved antimicrobials. The human ribosome is much more complex than its bacterial counterpart. We have started analyzing eukaryotic-specific steps of protein synthesis by the human ribosome to understand the highly regulated processes that control human gene expression.



